Code
knitr::include_graphics("figures/table_originalitems.png")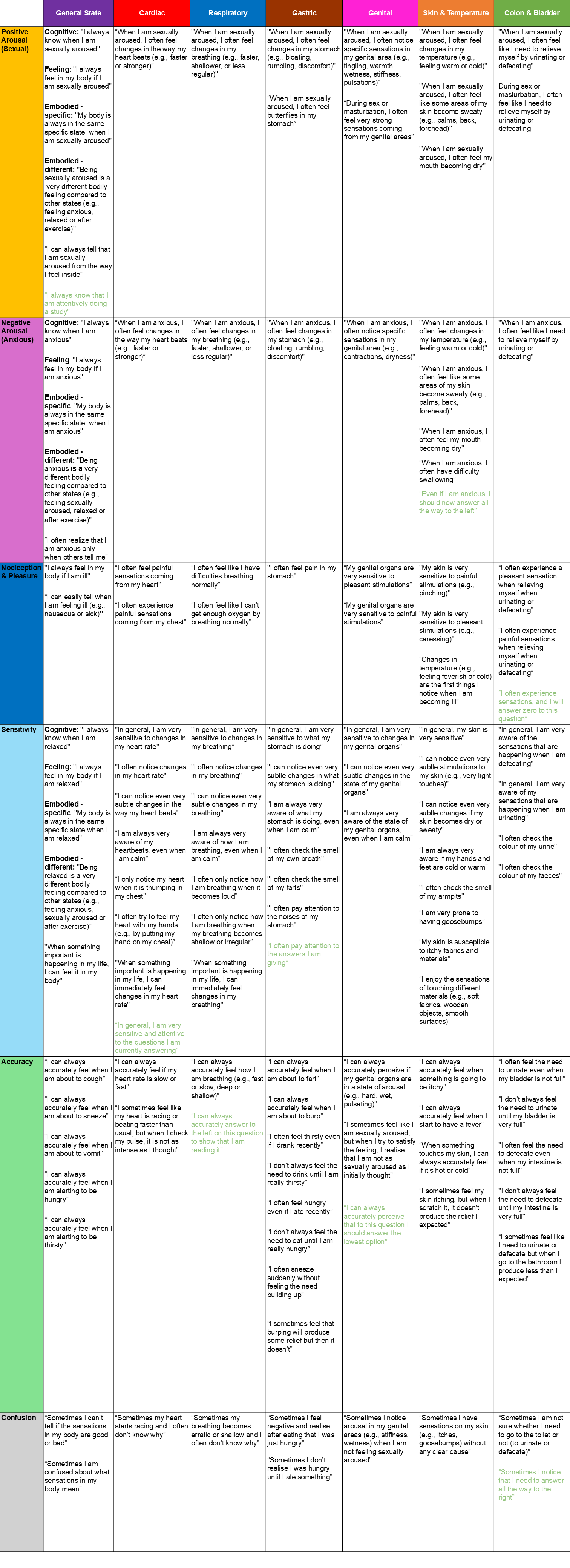
knitr::include_graphics("figures/table_originalitems.png")
library(tidyverse)
library(easystats)
library(patchwork)
library(ggside)
library(EGAnet)
library(tidygraph)
library(ggraph)
set.seed(42)df <- read.csv("../data/data_participants.csv")
items <- select(df, contains("_Q"))
byfacet <- items[df$Condition=="Dimensions",]
bymodality <- items[df$Condition=="Domains",]
byrandom <- items[df$Condition=="Random",]labels <- list(
Sexual_State_Q1 = "I always know when I am sexually aroused",
Sexual_State_Q2 = "I always feel in my body if I am sexually aroused",
Sexual_State_Q3 = "My body is always in the same specific state when I am sexually aroused",
Sexual_State_Q4 = "Being sexually aroused is a very different bodily feeling compared to other states (e.g., feeling anxious, relaxed, or after physical exercise)",
Sexual_State_Q5 = "I can always tell that I am sexually aroused from the way I feel inside",
Sexual_State_A = "I always know that I am attentively doing a study",
Sexual_Cardiac_Q1 = "When I am sexually aroused, I often feel changes in the way my heart beats (e.g., faster or stronger)",
Sexual_Respiratory_Q1 = "When I am sexually aroused, I often feel changes in my breathing (e.g., faster, shallower, or less regular)",
Sexual_Gastric_Q1 = "When I am sexually aroused, I often feel changes in my stomach (e.g., bloating, rumbling, discomfort)",
Sexual_Gastric_Q2 = "When I am sexually aroused, I often feel butterflies in my stomach",
Sexual_Genital_Q1 = "When I am sexually aroused, I often notice specific sensations in my genital area (e.g., tingling, warmth, wetness, stiffness, pulsations)",
Sexual_Genital_Q2 = "During sex or masturbation, I often feel very strong sensations coming from my genital areas",
Sexual_SkinThermo_Q1 = "When I am sexually aroused, I often feel changes in my temperature (e.g., feeling warm or cold)",
Sexual_SkinThermo_Q2 = "When I am sexually aroused, I often feel like some areas of my skin become sweaty (e.g., palms, back, forehead)",
Sexual_SkinThermo_Q3 = "When I am sexually aroused, I often feel my mouth becoming dry",
Sexual_ColonBladder_Q1 = "When I am sexually aroused, I often feel like I need to relieve myself by urinating or defecating",
Sexual_ColonBladder_Q2 = "During sex or masturbation, I often feel like I need to relieve myself by urinating or defecating",
Anxious_State_Q1 = "I always know when I am anxious",
Anxious_State_Q2 = "I always feel in my body if I am anxious",
Anxious_State_Q3 = "My body is always in the same specific state when I am anxious",
Anxious_State_Q4 = "Being anxious is a very different bodily feeling compared to other states (e.g., feeling sexually aroused, relaxed or after exercise)",
Anxious_State_Q5 = "I often realize that I am anxious only when others tell me",
Anxious_Cardiac_Q1 = "When I am anxious, I often feel changes in the way my heart beats (e.g., faster or stronger)",
Anxious_Respiratory_Q1 = "When I am anxious, I often feel changes in my breathing (e.g., faster, shallower, or less regular)",
Anxious_Gastric_Q1 = "When I am anxious, I often feel changes in my stomach (e.g., bloating, rumbling, discomfort)",
Anxious_Genital_Q1 = "When I am anxious, I often notice specific sensations in my genital area (e.g., contractions, dryness)",
Anxious_SkinThermo_Q1 = "When I am anxious, I often feel changes in my temperature (e.g., feeling warm or cold)",
Anxious_SkinThermo_Q2 = "When I am anxious, I often feel like some areas of my skin become sweaty (e.g., palms, back, forehead)",
Anxious_SkinThermo_Q3 = "When I am anxious, I often feel my mouth becoming dry",
Anxious_SkinThermo_Q4 = "When I am anxious, I often have difficulty swallowing",
Anxious_SkinThermo_A = "Even if I am anxious, I should now answer all the way to the left",
Anxious_ColonBladder_Q1 = "When I am anxious, I often feel like I need to relieve myself by urinating or defecating",
Nociception_State_Q1 = "I always feel in my body if I am ill",
Nociception_State_Q2 = "I can easily tell when I am feeling ill (e.g., nauseous or sick)",
Nociception_Cardiac_Q1 = "I often feel painful sensations coming from my heart",
Nociception_Cardiac_Q2 = "I often experience painful sensations coming from my chest",
Nociception_Respiratory_Q1 = "I often feel like I have difficulties breathing normally",
Nociception_Respiratory_Q2 = "I often feel like I can't get enough oxygen by breathing normally",
Nociception_Gastric_Q1 = "I often feel pain in my stomach",
Nociception_Genital_Q1 = "My genital organs are very sensitive to pleasant stimulations",
Nociception_Genital_Q2 = "My genital organs are very sensitive to painful stimulations",
Nociception_SkinThermo_Q1 = "My skin is very sensitive to painful stimulations (e.g., pinching)",
Nociception_SkinThermo_Q2 = "My skin is very sensitive to pleasant stimulations (e.g., caressing)",
Nociception_SkinThermo_Q3 = "Changes in temperature (e.g., feeling feverish or cold) are the first things I notice when I am becoming ill",
Nociception_ColonBladder_Q1 = "I often experience a pleasant sensation when relieving myself when urinating or defecating)",
Nociception_ColonBladder_Q2 = "I often experience painful sensations when relieving myself when urinating or defecating",
Nociception_ColonBladder_A = "I often experience sensations, and I will answer zero to this question",
Sensitivity_State_Q1 = "I always know when I am relaxed",
Sensitivity_State_Q2 = "I always feel in my body if I am relaxed",
Sensitivity_State_Q3 = "My body is always in the same specific state when I am relaxed",
Sensitivity_State_Q4 = "Being relaxed is a very different bodily feeling compared to other states (e.g., feeling anxious, sexually aroused or after exercise)",
Sensitivity_State_Q5 = "When something important is happening in my life, I can feel it in my body",
Sensitivity_Cardiac_Q1 = "In general, I am very sensitive to changes in my heart rate",
Sensitivity_Cardiac_Q2 = "I often notice changes in my heart rate",
Sensitivity_Cardiac_Q3 = "I can notice even very subtle changes in the way my heart beats",
Sensitivity_Cardiac_Q4 = "I am always very aware of my heartbeats, even when I am calm",
Sensitivity_Cardiac_Q5 = "I only notice my heart when it is thumping in my chest",
Sensitivity_Cardiac_Q6 = "I often try to feel my heart with my hands (e.g., by putting my hand on my chest)",
Sensitivity_Cardiac_Q7 = "When something important is happening in my life, I can feel immediately feel changes in my heart rate",
Sensitivity_Cardiac_A = "In general, I am very sensitive and attentive to the questions I am currently answering",
Sensitivity_Respiratory_Q1 = "In general, I am very sensitive to changes in my breathing",
Sensitivity_Respiratory_Q = "I often notice changes in my breathing",
Sensitivity_Respiratory_Q3 = "I can notice even very subtle changes in my breathing",
Sensitivity_Respiratory_Q4 = "I am always very aware of how I am breathing, even when I am calm",
Sensitivity_Respiratory_Q5 = "I often only notice how I am breathing when it becomes loud",
Sensitivity_Respiratory_Q6 = "I often only notice how I am breathing when my breathing becomes shallow or irregular",
Sensitivity_Respiratory_Q7 = "When something important is happening in my life, I can immediately feel changes in my breathing",
Sensitivity_Gastric_Q1 = "In general, I am very sensitive to what my stomach is doing",
Sensitivity_Gastric_Q2 = "I can notice even very subtle changes in what my stomach is doing",
Sensitivity_Gastric_Q3 = "I am always very aware of what my stomach is doing, even when I am calm",
Sensitivity_Gastric_Q4 = "I often check the smell of my own breath",
Sensitivity_Gastric_Q5 = "I often check the smell of my farts",
Sensitivity_Gastric_Q6 = "I often pay attention to the noises of my stomach",
Sensitivity_Gastric_A = "I often pay attention to the answers I am giving",
Sensitivity_Genital_Q1 = "In general, I am very sensitive to changes in my genital organs",
Sensitivity_Genital_Q2 = "I can notice even very subtle changes in the state of my genital organs",
Sensitivity_Genital_Q3 = "I am always very aware of the state of my genital organs, even when I am calm",
Sensitivity_SkinThermo_Q1 = "In general, my skin is very sensitive",
Sensitivity_SkinThermo_Q2 = "I can notice even very subtle stimulations to my skin (e.g., very light touches)",
Sensitivity_SkinThermo_Q3 = "I can notice even very subtle changes if my skin becomes dry or sweaty",
Sensitivity_SkinThermo_Q4 ="I am always very aware if my hands and feet are cold or warm",
Sensitivity_SkinThermo_Q5 = "I often check the smell of my armpits",
Sensitivity_SkinThermo_Q6 = "I am very prone to having goosebumps",
Sensitivity_SkinThermo_Q7 ="My skin is susceptible to itchy fabrics and materials",
Sensitivity_SkinThermo_Q8 = "I enjoy the sensations of touching different materials (e.g., soft fabrics, wooden objects, smooth surfaces)",
Sensitivity_ColonBladder_Q1 = "In general, I am very aware of the sensations that are happening when I am defecating",
Sensitivity_ColonBladder_Q2 = "In general, I am very aware of the sensations that are happening when I am urinating",
Sensitivity_ColonBladder_Q3 = "I often check the colour of my urine",
Sensitivity_ColonBladder_Q4 = "I often check the colour of my faeces",
Accuracy_State_Q1 = "I can always accurately feel when I am about to cough",
Accuracy_State_Q2 = "I can always accurately feel when I am about to sneeze",
Accuracy_State_Q3 = "I can always accurately feel when I am about to vomit",
Accuracy_State_Q4 = "I can always accurately feel when I am starting to be hungry",
Accuracy_State_Q5 = "I can always accurately feel when I am starting to be thirsty",
Accuracy_Cardiac_Q1 = "I can always accurately feel if my heart rate is slow or fast",
Accuracy_Cardiac_Q2 = "I sometimes feel like my heart is racing or beating faster than usual, but when I check my pulse, it is not as intense as I thought",
Accuracy_Respiratory_Q1 = "I can always accurately feel how I am breathing (e.g., fast or slow, deep or shallow)",
Accuracy_Respiratory_A = "I can always accurately answer to the left on this question to show that I am reading it",
Accuracy_Gastric_Q1 = "I can always accurately feel when I am about to fart",
Accuracy_Gastric_Q2 = "I can always accurately feel when I am about to burp",
Accuracy_Gastric_Q3 = "I often feel thirsty even if I drank recently",
Accuracy_Gastric_Q4 = "I don't always feel the need to drink until I am really thirsty",
Accuracy_Gastric_Q5 = "I often feel hungry even if I ate recently",
Accuracy_Gastric_Q6 = "I don't always feel the need to eat until I am really hungry",
Accuracy_Gastric_Q7 = "I often sneeze suddenly without feeling the need building up",
Accuracy_Gastric_Q8 = "I sometimes feel that burping will produce some relief but then it doesn't",
Accuracy_Genital_Q1 = "I can always accurately perceive if my genital organs are in a state of arousal (e.g., hard, wet, pulsating)",
Accuracy_Genital_Q2 = "I sometimes feel like I am sexually aroused, but when I try to satisfy the feeling, I realise that I am not as sexually aroused as I initially thought",
Accuracy_Genital_A = "I can always accurately perceive that to this question I should answer the lowest option",
Accuracy_SkinThermo_Q1 = "I can always accurately feel when something is going to be itchy",
Accuracy_SkinThermo_Q2 = "I can always accurately feel when I start to have a fever",
Accuracy_SkinThermo_Q3 = "When something touches my skin, I can always accurately feel if it's hot or cold",
Accuracy_SkinThermo_Q4 = "I sometimes feel my skin itching, but when I scratch it, it doesn't produce the relief I expected",
Accuracy_ColonBladder_Q1 = "I often feel the need to urinate even when my bladder is not full",
Accuracy_ColonBladder_Q2 = "I don't always feel the need to urinate until my bladder is very full",
Accuracy_ColonBladder_Q3 = "I often feel the need to defecate even when my intestine is not full",
Accuracy_ColonBladder_Q4 = "I don't always feel the need to defecate until my intestine is very full",
Accuracy_ColonBladder_Q5 = "I sometimes feel like I need to urinate or defecate but when I go to the bathroom I produce less than I expected",
Confusion_State_Q1 = "Sometimes I can't tell if the sensations in my body are good or bad",
Confusion_State_Q2 = "Sometimes I am confused about what sensations in my body mean",
Confusion_Cardiac_Q1 = "Sometimes my heart starts racing and I often don't know why",
Confusion_Respiratory_Q1 = "Sometimes my breathing becomes erratic or shallow and I often don't know why",
Confusion_Gastric_Q1 = "Sometimes I feel negative and realise after eating that I was just hungry",
Confusion_Gastric_Q2 = "Sometimes I don't realise I was hungry until I ate something",
Confusion_Genital_Q1 = "Sometimes I notice arousal in my genital areas (e.g., stiffness, wetness) when I am not feeling sexually aroused",
Confusion_SkinThermo_Q1 = "Sometimes I have sensations on my skin (e.g., itches, goosebumps) without any clear cause",
Confusion_ColonBladder_Q1 = "Sometimes I am not sure whether I need to go to the toilet or not (to urinate or defecate)",
Confusion_ColonBladder_A = "Sometimes I notice that I need to answer all the way to the right"
)make_heatmap <- function(items, interactive=TRUE, title="Correlation Matrix") {
fn <- if(interactive) heatmaply::heatmaply else heatmaply::ggheatmap
fn(
cor(items),
symm = TRUE,
colors=heatmaply::cool_warm,
show_dendrogram =c(FALSE, TRUE),
main=title,
showticklabels = FALSE,
hide_colorbar = TRUE,
k_col = 4,
# k_row = 4,
limits = c(-1, 1)
)
}
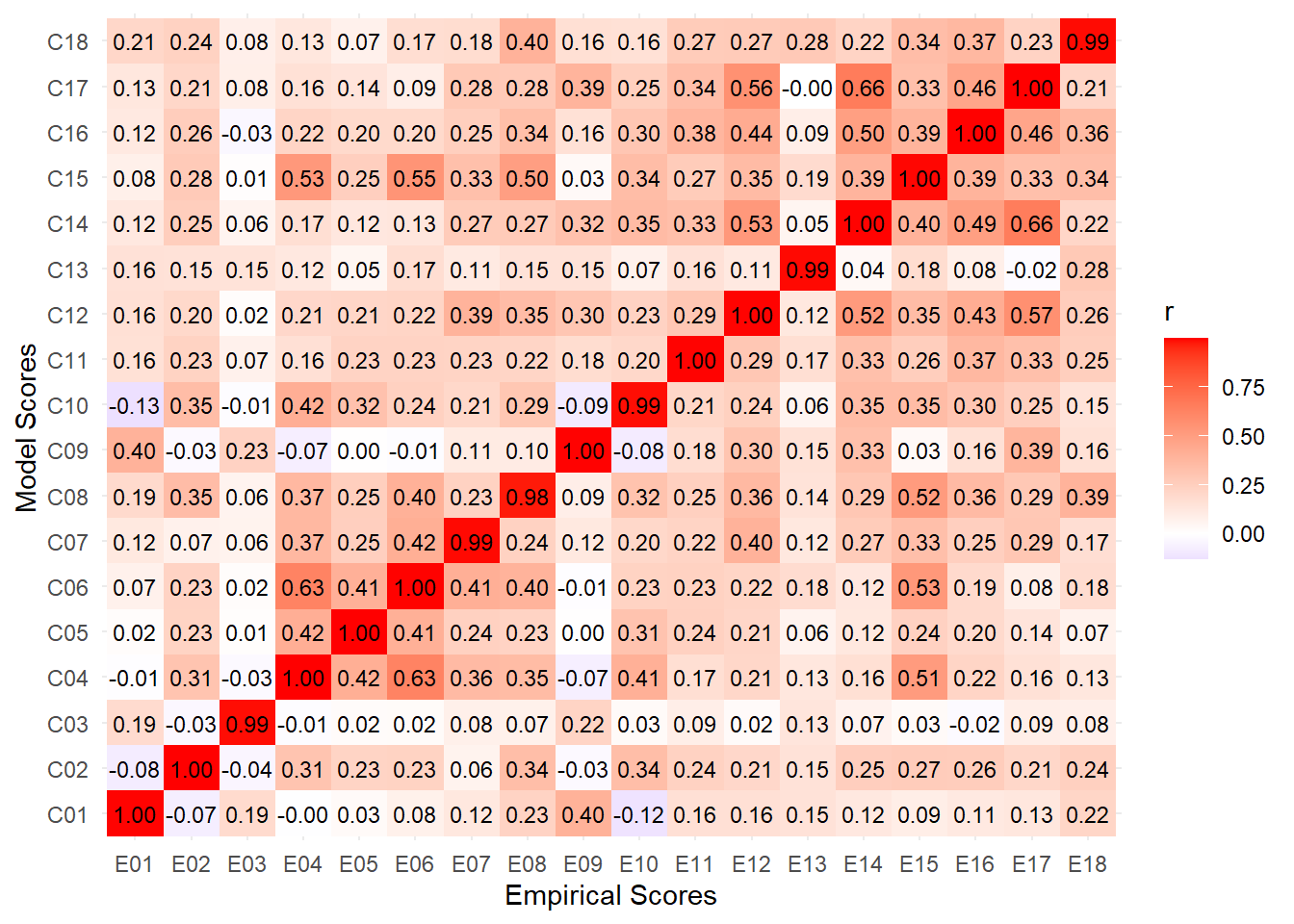
make_heatmap(items, title="Full Sample")bootstrap_similarity <- function(df) {
rez <- data.frame()
for(i in 1:500) {
idx <- sample(1:nrow(df), nrow(df), replace=TRUE)
newdata <- df[idx,]
items <- select(df, contains("_Q"))
byfacet <- items[newdata$Condition=="Dimensions",]
bymodality <- items[newdata$Condition=="Domains",]
byrandom <- items[newdata$Condition=="Random",]
rez <- rbind(
MatrixCorrelation::allCorrelations(cor(byrandom), cor(byfacet),
ncomp1=10, ncomp2=10, plot=FALSE),
MatrixCorrelation::allCorrelations(cor(byrandom), cor(bymodality),
ncomp1=10, ncomp2=10, plot=FALSE),
MatrixCorrelation::allCorrelations(cor(items), cor(byfacet),
ncomp1=10, ncomp2=10, plot=FALSE),
MatrixCorrelation::allCorrelations(cor(items), cor(bymodality),
ncomp1=10, ncomp2=10, plot=FALSE)) |>
as.data.frame() |>
mutate(
Comparison = c("Random vs. Facet", "Random vs. Modality", "Full vs. Facet", "Full vs. Modality"),
Sample = c("Random", "Random", "Full", "Full"),
Iteration = i) |>
mutate(d_PSI = diff(PSI), d_RVadj = diff(RVadj), d_SMI = diff(SMI), .by="Sample") |>
rbind(rez)
}
rez
}
rez <- bootstrap_similarity(df)rez |>
summarize(PSI = mean(PSI),
p_PSI = as.numeric(bayestestR::pd(d_PSI, as_p=TRUE)),
RVadj = mean(RVadj),
p_RVadj = as.numeric(bayestestR::pd(d_RVadj, as_p=TRUE)),
SMI = mean(SMI),
p_SMI = as.numeric(bayestestR::pd(d_SMI, as_p=TRUE)),
.by = "Comparison") Comparison PSI p_PSI RVadj p_RVadj SMI p_SMI
1 Random vs. Facet 0.810526 0.448 0.768536 0.744 0.493086 0.524
2 Random vs. Modality 0.822352 0.448 0.784162 0.744 0.512444 0.524
3 Full vs. Facet 0.925638 0.212 0.908974 0.444 0.724340 0.436
4 Full vs. Modality 0.941368 0.212 0.929352 0.444 0.764880 0.436rez |>
pivot_longer(-c("Comparison", "Sample", "Iteration")) |>
filter(name %in% c("PSI", "RVadj", "SMI")) |>
mutate(side = ifelse(str_detect(Comparison, "Facet"), "left", "right")) |>
ggplot(aes(x=name, y=value, fill=Comparison, color=Comparison)) +
ggdist::stat_halfeye(aes(side=side), position=position_dodge(width=0.3), alpha=0.7) +
facet_wrap(~Sample) +
scale_fill_manual(values=c("Random vs. Facet"="#F44336", "Random vs. Modality"="#2196F3", "Full vs. Facet"="#FF5722", "Full vs. Modality"="#00BCD4")) +
scale_color_manual(values=c("Random vs. Facet"="#F44336", "Random vs. Modality"="#2196F3", "Full vs. Facet"="#FF5722", "Full vs. Modality"="#00BCD4")) +
labs(x="Indices of Correlation Matrix Difference", y="Similarity") +
theme_minimal()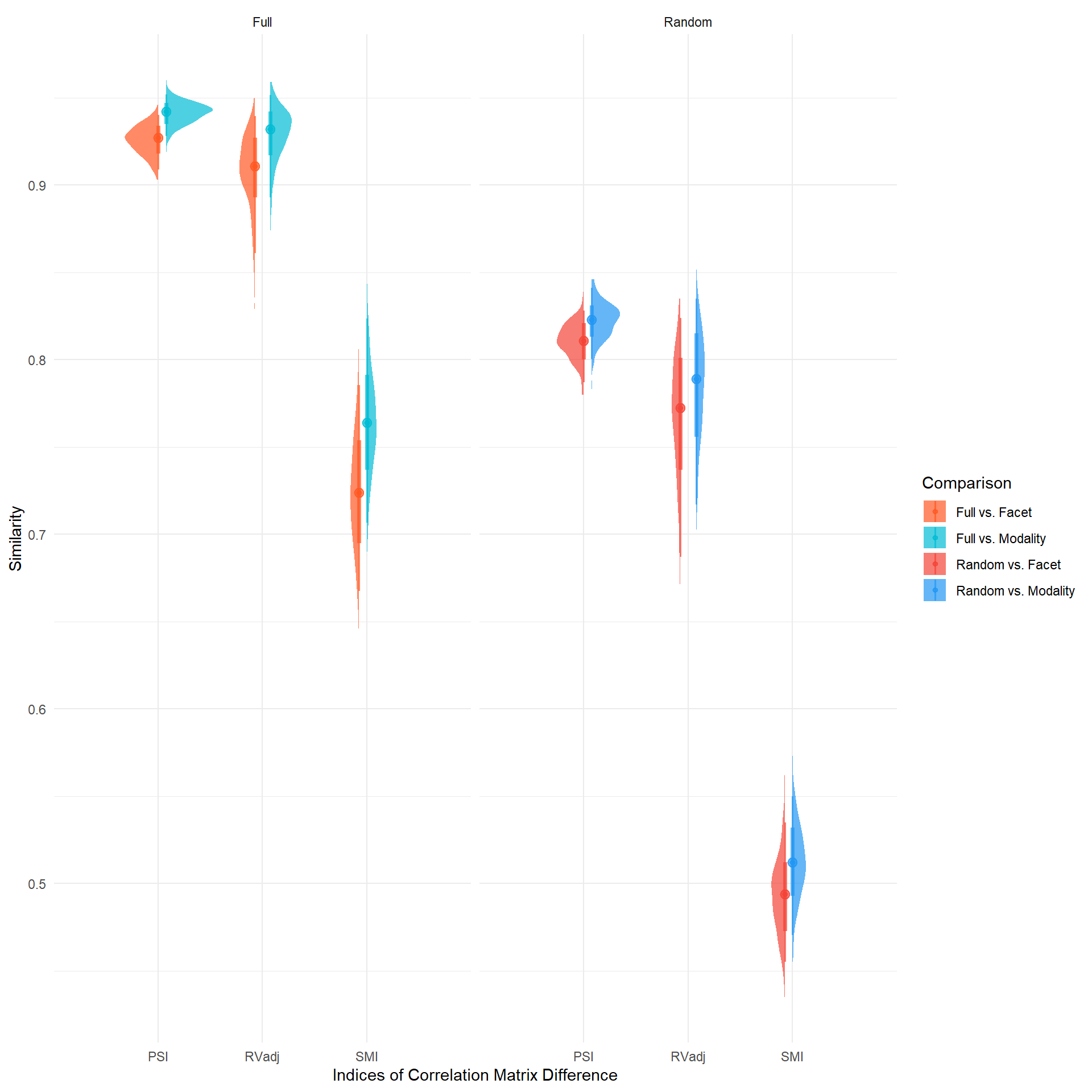
p_sim <- rez |>
pivot_longer(-c("Comparison", "Sample", "Iteration")) |>
filter(str_detect(name, "d_") & str_detect(Comparison, "Facet")) |>
filter(Sample == "Random") |>
mutate(name = str_remove(name, "d_"), value = -value) |> # Reverse score for figure aesthetics
ggplot(aes(x=value, y=name)) +
ggdist::stat_eye(aes(fill = after_stat(x < 0))) +
geom_vline(xintercept=0, linetype="dashed") +
labs(x="Similarity Difference", y="Indices of pattern difference",
title="Which matrix is the above more similar to?",
fill="More similar to") +
theme_minimal() +
scale_fill_manual(values=c("TRUE"="#4CAF50", "FALSE"="#2196F3"),
labels = c("By facet", "By modality"),
guide = guide_legend(reverse = TRUE)) +
theme(plot.title = element_text(hjust = 0.5, face="italic"))
r <- correlation::cor_sort(cor(byrandom))
order <- colnames(r)
make_corplot <- function(data, order) {
rownames_to_column(as.data.frame(cor(data)), "item") |>
pivot_longer(-item) |>
mutate(item = fct_relevel(item, order),
name = fct_relevel(name, order)) |>
ggplot(aes(x=name, y=item, fill=value)) +
geom_tile() +
# scale_fill_gradient2(low="blue", high="red", mid="white", midpoint=0, values=c(-1, 0, 1)) +
scale_fill_gradientn(
colors=c("blue", "white", "#FFEE58", "#FFC107", "#FF9800", "#FF5722",
"#F44336", "#E91E63", "#9C27B0", "#8231B4", "#673AB7", "white"),
values=c(-1, seq(0, 1, 0.1))) +
theme_minimal() +
theme(axis.text = element_blank(),
axis.title = element_blank(),
legend.position = "none")
}
p1 <- make_corplot(byrandom, order) +
ggtitle("Grouped randomly")
p2 <- make_corplot(byfacet, order) +
ggtitle("Grouped by facet")
p3 <- make_corplot(bymodality, order) +
ggtitle("Grouped by modality")
fig1 <- p1 / (p3 | p_sim | p2) +
patchwork::plot_layout(heights=c(3, 1)) +
patchwork::plot_annotation(
title="Item Correlation Matrix Similarity",
theme = theme(plot.title = element_text(hjust = 0.5, face="bold")))
fig1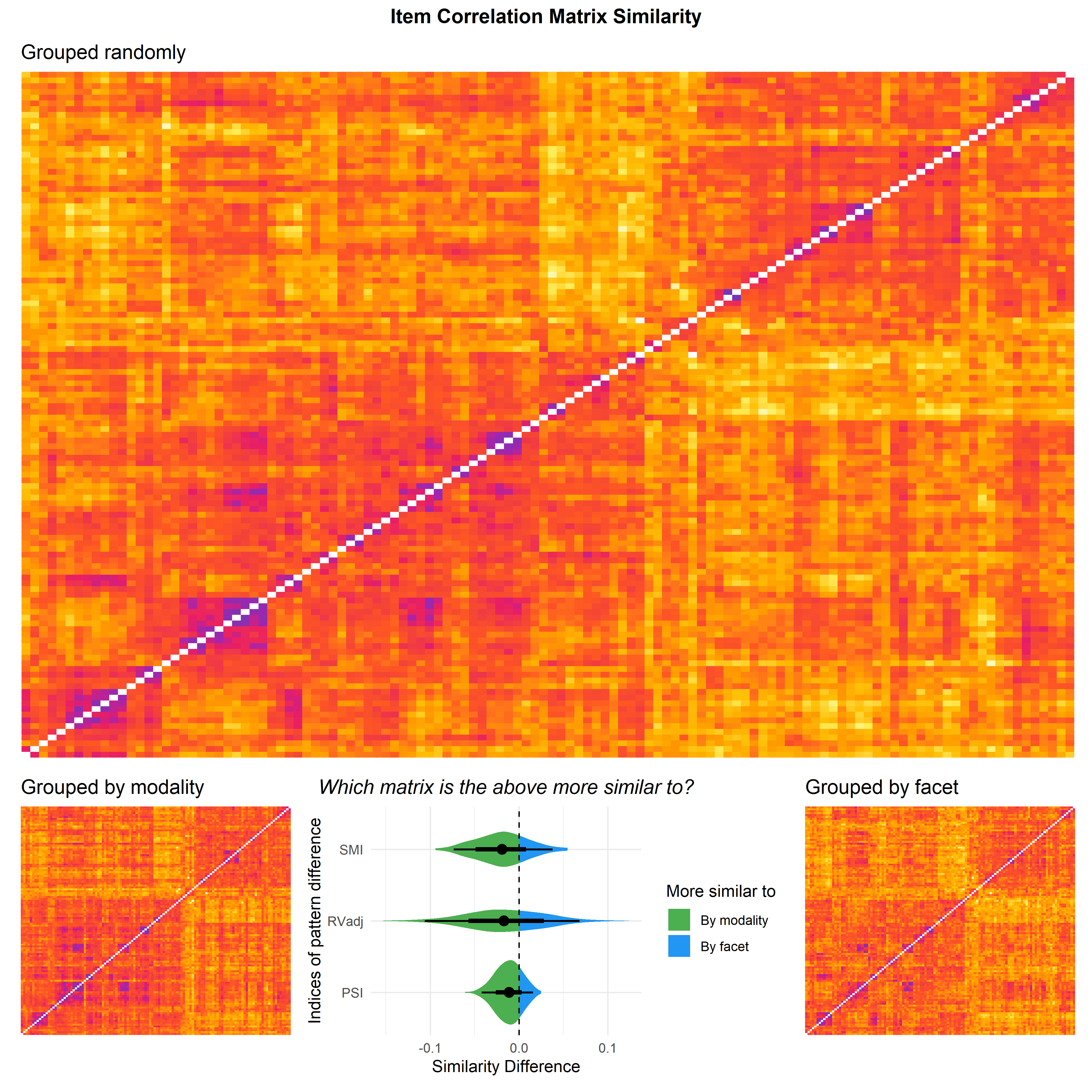
ggsave("figures/fig1_correlations.png", fig1, scale = 1.25, width=2100, height=2100, dpi=300, units = "px")items |>
bayestestR::estimate_density(method="kernSmooth") |>
separate(Parameter, into=c("Facet", "Modality", "Item"), sep="_") |>
ggplot(aes(x=x, y=y)) +
geom_line(aes(color=Facet, group=interaction(Facet, Item)), linewidth=1, alpha=0.9) +
facet_wrap(~Modality) +
theme_minimal() +
scale_color_material_d(palette = "rainbow") +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank())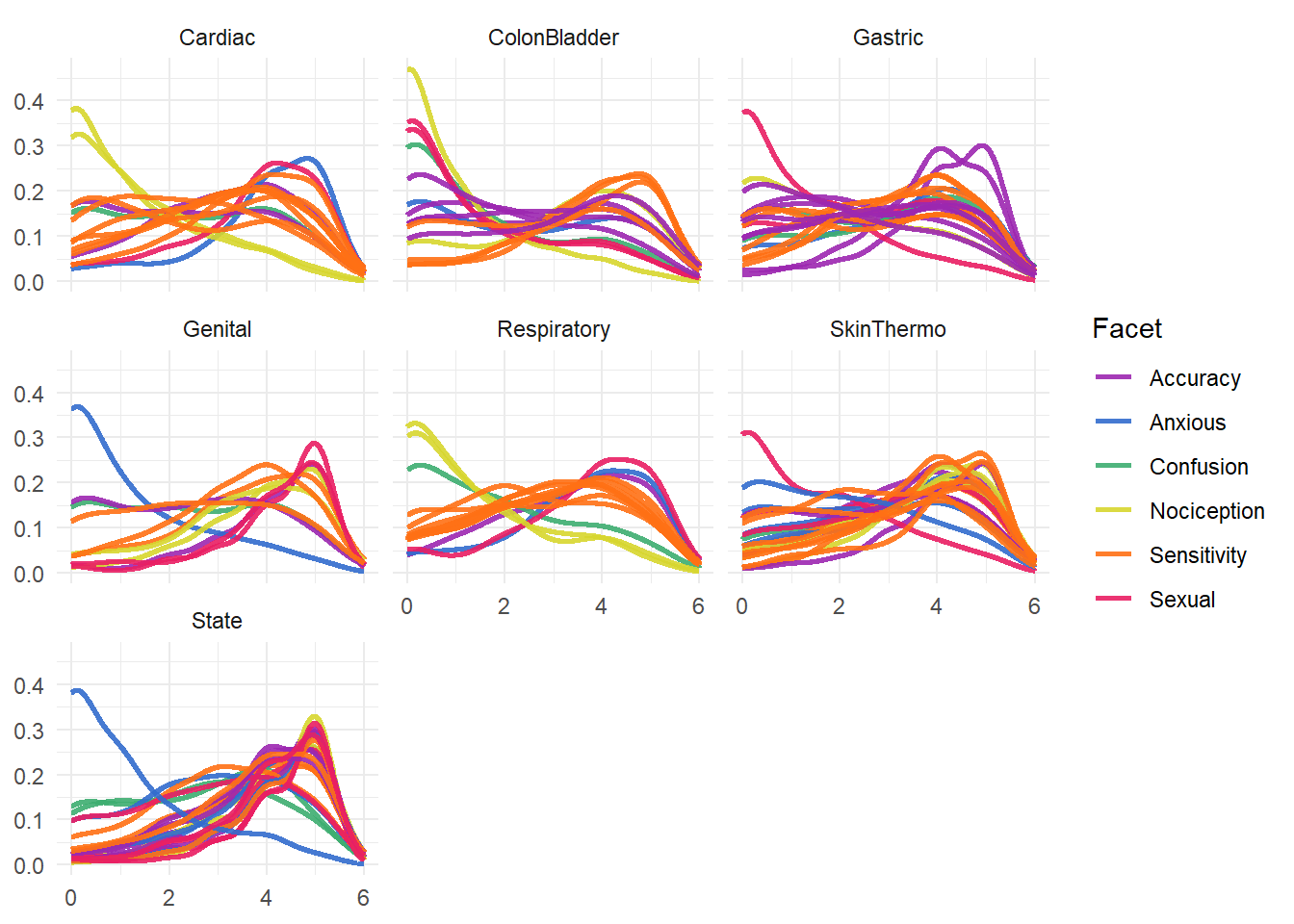
uva <- EGAnet::UVA(items, cut.off = 0.25, reduce.method="remove", verbose=FALSE)
toremove1 <- unique(uva$keep_remove$remove)
keep <- uva$keep_remove$keep[!uva$keep_remove$keep %in% toremove1]
sorted <- correlation::cor_sort(cor(items[toremove1],items[keep]))
correlation::correlation(items[toremove1], items[keep]) |>
mutate(Parameter1 = fct_relevel(Parameter1, rownames(sorted)),
Parameter2 = fct_relevel(Parameter2, colnames(sorted))) |>
ggplot(aes(x=Parameter1, y=Parameter2, fill=r)) +
geom_tile() +
geom_text(aes(label=insight::format_value(r)), color="white") +
scale_fill_gradient2(low="blue", high="red", mid="white", midpoint=0, guide="none") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
labs(x="Removed Items", y="Kept Items", fill="Correlation")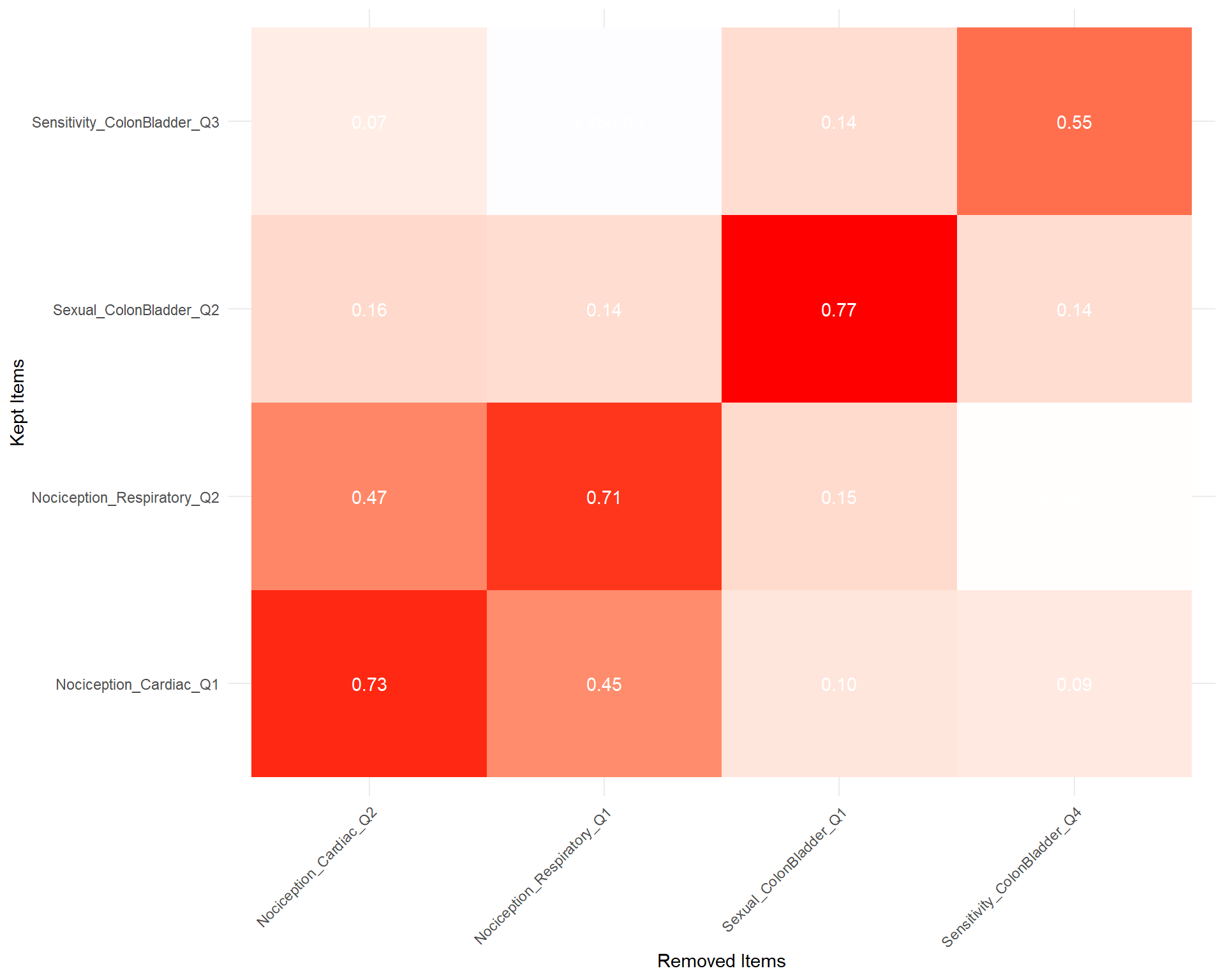
items <- select(items, -all_of(toremove1))
byfacet <- select(byfacet, -all_of(toremove1))
bymodality <- select(bymodality, -all_of(toremove1))
byrandom <- select(byrandom, -all_of(toremove1))We removed 4 items (Sexual_ColonBladder_Q1, Nociception_Respiratory_Q1, Nociception_Cardiac_Q2, Sensitivity_ColonBladder_Q4) based on Unique Variable Analysis (UVA).
ega1 <- bootEGA(
items,
EGA.type = "hierEGA",
model = "glasso", # BGGM
algorithm = "leiden",
type="resampling",
plot.itemStability=FALSE,
typicalStructure=TRUE,
plot.typicalGraph=FALSE,
iter=500,
seed=3, ncores = 4)
save(ega1, file="models/ega1.RData")load("models/ega1.RData")
# EGAnet::dimensionStability(ega)
itemstability <- EGAnet::itemStability(ega1, IS.plot=FALSE)
plot(itemstability)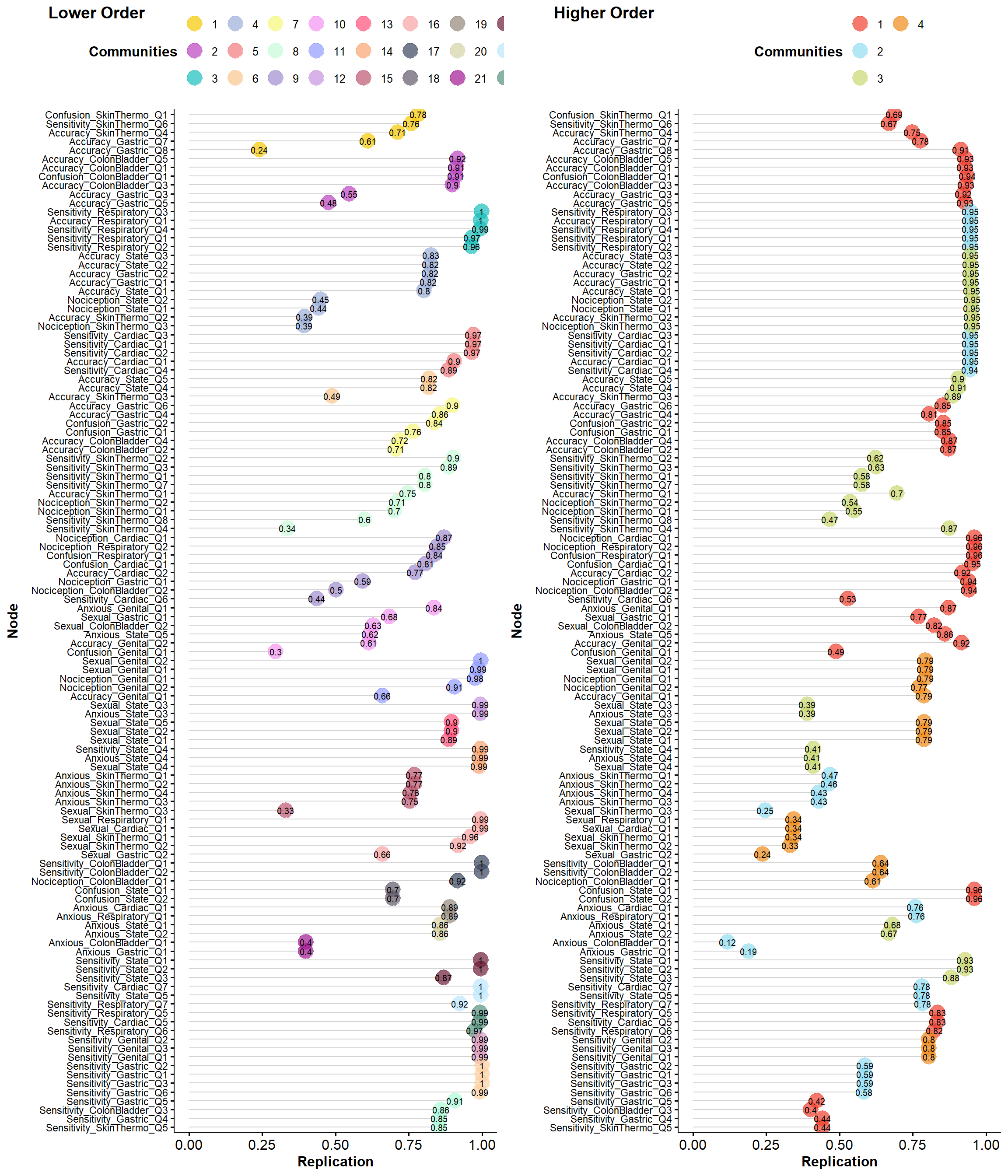
make_loadingtable <- function(x) {
t <- as.data.frame(x) |>
datawizard::data_addprefix("C")
t$Cluster <- colnames(t)[max.col(t, ties.method='first')]
t |>
rownames_to_column(var="Item") |>
rowwise() |>
mutate(Max = max(c_across(-c(Item, Cluster)))) |>
arrange(Cluster, desc(Max)) |>
as.data.frame()
}
itemstability_table <- make_loadingtable(itemstability$lower_order$item.stability$all.dimensions)
# Remove items
toremove2 <- filter(itemstability_table, Max < 0.8)$ItemWe removed 40 items (Confusion_SkinThermo_Q1, Sensitivity_SkinThermo_Q6, Accuracy_SkinThermo_Q4, Accuracy_Gastric_Q7, Accuracy_Gastric_Q8, Accuracy_Gastric_Q3, Accuracy_Gastric_Q5, Nociception_State_Q2, Nociception_State_Q1, Accuracy_SkinThermo_Q2, Nociception_SkinThermo_Q3, Accuracy_SkinThermo_Q3, Confusion_Gastric_Q1, Accuracy_ColonBladder_Q4, Accuracy_ColonBladder_Q2, Accuracy_SkinThermo_Q1, Nociception_SkinThermo_Q2, Nociception_SkinThermo_Q1, Sensitivity_SkinThermo_Q8, Sensitivity_SkinThermo_Q4, Accuracy_Cardiac_Q2, Nociception_Gastric_Q1, Nociception_ColonBladder_Q2, Sensitivity_Cardiac_Q6, Sexual_Gastric_Q1, Sexual_ColonBladder_Q2, Anxious_State_Q5, Accuracy_Genital_Q2, Accuracy_Genital_Q1, Anxious_SkinThermo_Q1, Anxious_SkinThermo_Q2, Anxious_SkinThermo_Q4, Anxious_SkinThermo_Q3, Sexual_SkinThermo_Q3, Sexual_Gastric_Q2, Confusion_State_Q1, Confusion_State_Q2, Anxious_ColonBladder_Q1, Anxious_Gastric_Q1, Confusion_Genital_Q1) that had a low stability (< 0.8).
toremove2b <- itemstability_table |>
filter(!Item %in% toremove2) |>
mutate(Item, n=n(), .by="Cluster", .keep="used") |>
filter(n < 3) |> # Remove small clusters
pull(Item)
toremove2 <- c(toremove2, toremove2b)We removed 9 items (Accuracy_State_Q5, Accuracy_State_Q4, Anxious_Genital_Q1, Sexual_State_Q3, Anxious_State_Q3, Anxious_Cardiac_Q1, Anxious_Respiratory_Q1, Anxious_State_Q1, Anxious_State_Q2) that were in clusters with less than 3 items after the first removal.
# plot(itemstability)itemstability_table |>
select(-Cluster, -Max) |>
gt::gt() |>
gt::data_color(columns=-Item,
method = "numeric",
palette = c("white", "orange", "green"),
domain = c(0, 1)) |>
gt::data_color(columns=Item, fn=\(x) ifelse(x %in% toremove2, "#FFEBEE", "white")) |>
gt::tab_header(
title = gt::md("**Item Stability**"),
subtitle = "Proportion of times an item is assigned to the same cluster"
) |>
gt::fmt_number(columns=-Item, decimals=3)| Item Stability | ||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proportion of times an item is assigned to the same cluster | ||||||||||||||||||||||||||||||||||
| Item | C01 | C02 | C03 | C04 | C05 | C06 | C07 | C08 | C09 | C10 | C11 | C12 | C13 | C14 | C15 | C16 | C17 | C18 | C19 | C20 | C21 | C22 | C23 | C24 | C25 | C26 | C27 | C28 | C29 | C30 | C31 | C32 | C33 | C34 |
| Confusion_SkinThermo_Q1 | 0.782 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.182 | 0.010 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.002 | 0.000 | 0.006 | 0.004 | 0.002 | 0.000 |
| Sensitivity_SkinThermo_Q6 | 0.758 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.182 | 0.010 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.002 | 0.006 | 0.008 | 0.000 | 0.006 | 0.004 | 0.002 | 0.000 |
| Accuracy_SkinThermo_Q4 | 0.712 | 0.046 | 0.000 | 0.000 | 0.000 | 0.000 | 0.014 | 0.138 | 0.014 | 0.030 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.012 | 0.022 | 0.006 | 0.002 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q7 | 0.610 | 0.148 | 0.000 | 0.004 | 0.004 | 0.000 | 0.004 | 0.068 | 0.012 | 0.082 | 0.000 | 0.000 | 0.000 | 0.004 | 0.002 | 0.004 | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.008 | 0.000 | 0.008 | 0.012 | 0.014 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q8 | 0.240 | 0.210 | 0.000 | 0.000 | 0.000 | 0.000 | 0.012 | 0.016 | 0.108 | 0.112 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.050 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.124 | 0.000 | 0.000 | 0.038 | 0.024 | 0.042 | 0.012 | 0.004 | 0.000 | 0.000 | 0.000 |
| Accuracy_ColonBladder_Q5 | 0.002 | 0.916 | 0.000 | 0.000 | 0.000 | 0.000 | 0.024 | 0.000 | 0.014 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.026 | 0.012 | 0.000 | 0.002 | 0.000 | 0.000 |
| Accuracy_ColonBladder_Q1 | 0.000 | 0.912 | 0.000 | 0.000 | 0.000 | 0.000 | 0.032 | 0.000 | 0.014 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.024 | 0.010 | 0.000 | 0.002 | 0.000 | 0.000 |
| Confusion_ColonBladder_Q1 | 0.000 | 0.910 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.000 | 0.052 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.012 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 |
| Accuracy_ColonBladder_Q3 | 0.000 | 0.898 | 0.000 | 0.000 | 0.000 | 0.000 | 0.012 | 0.000 | 0.042 | 0.018 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.014 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q3 | 0.014 | 0.546 | 0.000 | 0.000 | 0.000 | 0.000 | 0.014 | 0.000 | 0.180 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.070 | 0.088 | 0.058 | 0.006 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q5 | 0.026 | 0.476 | 0.000 | 0.000 | 0.000 | 0.000 | 0.092 | 0.000 | 0.166 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.068 | 0.086 | 0.052 | 0.006 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Respiratory_Q3 | 0.000 | 0.000 | 0.998 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 |
| Accuracy_Respiratory_Q1 | 0.000 | 0.000 | 0.996 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Respiratory_Q4 | 0.000 | 0.000 | 0.994 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 |
| Sensitivity_Respiratory_Q1 | 0.000 | 0.000 | 0.966 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.002 | 0.002 | 0.006 | 0.004 | 0.004 |
| Sensitivity_Respiratory_Q2 | 0.000 | 0.000 | 0.964 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.002 | 0.002 | 0.006 | 0.004 | 0.004 |
| Accuracy_State_Q3 | 0.000 | 0.000 | 0.000 | 0.826 | 0.000 | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.112 | 0.046 | 0.004 | 0.002 | 0.000 | 0.000 | 0.000 |
| Accuracy_State_Q2 | 0.000 | 0.000 | 0.000 | 0.824 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.134 | 0.038 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q2 | 0.000 | 0.000 | 0.000 | 0.822 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.134 | 0.040 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q1 | 0.000 | 0.000 | 0.000 | 0.816 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.136 | 0.040 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_State_Q1 | 0.000 | 0.000 | 0.000 | 0.802 | 0.000 | 0.032 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.132 | 0.032 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 |
| Nociception_State_Q2 | 0.000 | 0.000 | 0.000 | 0.450 | 0.000 | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.336 | 0.084 | 0.060 | 0.040 | 0.020 | 0.002 | 0.000 |
| Nociception_State_Q1 | 0.000 | 0.000 | 0.000 | 0.442 | 0.000 | 0.002 | 0.000 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.336 | 0.080 | 0.058 | 0.038 | 0.020 | 0.002 | 0.000 |
| Accuracy_SkinThermo_Q2 | 0.000 | 0.000 | 0.000 | 0.394 | 0.000 | 0.000 | 0.000 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.376 | 0.148 | 0.038 | 0.022 | 0.006 | 0.000 | 0.000 |
| Nociception_SkinThermo_Q3 | 0.000 | 0.000 | 0.000 | 0.392 | 0.000 | 0.002 | 0.000 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.376 | 0.148 | 0.038 | 0.022 | 0.006 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q3 | 0.000 | 0.000 | 0.028 | 0.000 | 0.970 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q1 | 0.000 | 0.000 | 0.028 | 0.000 | 0.970 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q2 | 0.000 | 0.000 | 0.034 | 0.000 | 0.966 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_Cardiac_Q1 | 0.000 | 0.000 | 0.094 | 0.000 | 0.904 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q4 | 0.000 | 0.000 | 0.106 | 0.000 | 0.886 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_State_Q5 | 0.000 | 0.000 | 0.000 | 0.112 | 0.000 | 0.820 | 0.036 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.024 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_State_Q4 | 0.000 | 0.000 | 0.000 | 0.114 | 0.000 | 0.818 | 0.036 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.026 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_SkinThermo_Q3 | 0.000 | 0.000 | 0.000 | 0.182 | 0.000 | 0.488 | 0.008 | 0.136 | 0.008 | 0.000 | 0.010 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.084 | 0.058 | 0.012 | 0.004 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q6 | 0.000 | 0.030 | 0.000 | 0.000 | 0.000 | 0.006 | 0.898 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.038 | 0.024 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q4 | 0.000 | 0.060 | 0.000 | 0.000 | 0.000 | 0.072 | 0.856 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.006 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 |
| Confusion_Gastric_Q2 | 0.002 | 0.026 | 0.000 | 0.000 | 0.000 | 0.002 | 0.838 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.046 | 0.034 | 0.022 | 0.016 | 0.008 | 0.002 | 0.000 |
| Confusion_Gastric_Q1 | 0.022 | 0.020 | 0.000 | 0.000 | 0.000 | 0.000 | 0.764 | 0.016 | 0.034 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.046 | 0.034 | 0.018 | 0.018 | 0.008 | 0.002 | 0.000 |
| Accuracy_ColonBladder_Q4 | 0.000 | 0.176 | 0.000 | 0.000 | 0.000 | 0.000 | 0.720 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.022 | 0.042 | 0.032 | 0.006 | 0.000 | 0.000 | 0.000 |
| Accuracy_ColonBladder_Q2 | 0.000 | 0.184 | 0.000 | 0.000 | 0.000 | 0.002 | 0.706 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.022 | 0.044 | 0.034 | 0.006 | 0.000 | 0.000 | 0.000 |
| Sensitivity_SkinThermo_Q2 | 0.000 | 0.000 | 0.000 | 0.018 | 0.000 | 0.006 | 0.000 | 0.902 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.030 | 0.022 | 0.002 | 0.010 | 0.002 | 0.004 | 0.004 |
| Sensitivity_SkinThermo_Q3 | 0.000 | 0.000 | 0.000 | 0.026 | 0.000 | 0.002 | 0.000 | 0.886 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.038 | 0.022 | 0.004 | 0.010 | 0.002 | 0.004 | 0.004 |
| Sensitivity_SkinThermo_Q1 | 0.020 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.804 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.054 | 0.026 | 0.026 | 0.040 | 0.016 | 0.008 | 0.002 |
| Sensitivity_SkinThermo_Q7 | 0.020 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.804 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.054 | 0.026 | 0.026 | 0.040 | 0.016 | 0.008 | 0.002 |
| Accuracy_SkinThermo_Q1 | 0.002 | 0.000 | 0.000 | 0.144 | 0.000 | 0.002 | 0.000 | 0.746 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.070 | 0.032 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 |
| Nociception_SkinThermo_Q2 | 0.002 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.002 | 0.710 | 0.000 | 0.000 | 0.098 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.062 | 0.062 | 0.020 | 0.020 | 0.008 | 0.004 | 0.002 |
| Nociception_SkinThermo_Q1 | 0.030 | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.700 | 0.000 | 0.000 | 0.100 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.096 | 0.052 | 0.006 | 0.006 | 0.002 | 0.000 | 0.000 |
| Sensitivity_SkinThermo_Q8 | 0.086 | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.008 | 0.598 | 0.000 | 0.000 | 0.060 | 0.000 | 0.000 | 0.026 | 0.002 | 0.008 | 0.044 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.004 | 0.056 | 0.048 | 0.016 | 0.018 | 0.006 | 0.004 | 0.002 |
| Sensitivity_SkinThermo_Q4 | 0.002 | 0.000 | 0.000 | 0.174 | 0.000 | 0.218 | 0.000 | 0.336 | 0.000 | 0.000 | 0.004 | 0.002 | 0.000 | 0.004 | 0.000 | 0.002 | 0.012 | 0.002 | 0.000 | 0.018 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.152 | 0.056 | 0.016 | 0.002 | 0.000 | 0.000 | 0.000 |
| Nociception_Cardiac_Q1 | 0.000 | 0.022 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.870 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.052 | 0.036 | 0.012 | 0.006 | 0.000 | 0.000 | 0.000 |
| Nociception_Respiratory_Q2 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.848 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.022 | 0.024 | 0.048 | 0.028 | 0.024 | 0.004 | 0.000 |
| Confusion_Respiratory_Q1 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.838 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.020 | 0.022 | 0.048 | 0.030 | 0.024 | 0.004 | 0.000 |
| Confusion_Cardiac_Q1 | 0.002 | 0.000 | 0.000 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.808 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.034 | 0.060 | 0.062 | 0.022 | 0.002 | 0.000 | 0.000 |
| Accuracy_Cardiac_Q2 | 0.002 | 0.000 | 0.000 | 0.000 | 0.046 | 0.000 | 0.000 | 0.000 | 0.772 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.030 | 0.060 | 0.062 | 0.022 | 0.002 | 0.000 | 0.000 |
| Nociception_Gastric_Q1 | 0.004 | 0.178 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.592 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.024 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.000 | 0.074 | 0.062 | 0.030 | 0.010 | 0.006 | 0.000 | 0.000 |
| Nociception_ColonBladder_Q2 | 0.006 | 0.228 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.502 | 0.084 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.076 | 0.066 | 0.016 | 0.004 | 0.006 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q6 | 0.004 | 0.006 | 0.004 | 0.000 | 0.364 | 0.000 | 0.000 | 0.000 | 0.436 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.144 | 0.018 | 0.014 | 0.002 | 0.004 | 0.000 | 0.000 | 0.000 |
| Anxious_Genital_Q1 | 0.008 | 0.024 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.028 | 0.836 | 0.000 | 0.000 | 0.000 | 0.002 | 0.022 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.004 | 0.000 | 0.000 | 0.002 | 0.018 | 0.022 | 0.014 | 0.008 | 0.006 | 0.000 |
| Sexual_Gastric_Q1 | 0.002 | 0.052 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.684 | 0.000 | 0.000 | 0.000 | 0.000 | 0.016 | 0.056 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.034 | 0.068 | 0.050 | 0.030 | 0.000 | 0.000 | 0.000 |
| Sexual_ColonBladder_Q2 | 0.004 | 0.140 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.012 | 0.628 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.028 | 0.000 | 0.000 | 0.000 | 0.000 | 0.020 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.032 | 0.056 | 0.040 | 0.028 | 0.000 | 0.000 | 0.000 |
| Anxious_State_Q5 | 0.038 | 0.026 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.084 | 0.618 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.040 | 0.008 | 0.066 | 0.000 | 0.000 | 0.002 | 0.024 | 0.000 | 0.000 | 0.000 | 0.014 | 0.028 | 0.020 | 0.016 | 0.008 | 0.006 | 0.000 |
| Accuracy_Genital_Q2 | 0.204 | 0.032 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.052 | 0.614 | 0.000 | 0.002 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.004 | 0.000 | 0.000 | 0.010 | 0.032 | 0.026 | 0.006 | 0.000 | 0.000 | 0.000 |
| Sexual_Genital_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.996 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 |
| Sexual_Genital_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.986 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 |
| Nociception_Genital_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.976 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.010 | 0.004 | 0.000 | 0.002 | 0.002 | 0.000 |
| Nociception_Genital_Q2 | 0.002 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.022 | 0.000 | 0.000 | 0.906 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.024 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.024 | 0.004 | 0.002 | 0.002 | 0.002 | 0.000 |
| Accuracy_Genital_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.660 | 0.000 | 0.340 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sexual_State_Q3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.994 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_State_Q3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.994 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sexual_State_Q5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.104 | 0.000 | 0.896 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sexual_State_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.104 | 0.000 | 0.896 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sexual_State_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.114 | 0.000 | 0.886 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_State_Q4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.994 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_State_Q4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.992 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sexual_State_Q4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.006 | 0.990 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_SkinThermo_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.768 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.056 | 0.048 | 0.054 | 0.038 | 0.008 | 0.004 |
| Anxious_SkinThermo_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.768 | 0.012 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.056 | 0.048 | 0.054 | 0.038 | 0.008 | 0.004 |
| Anxious_SkinThermo_Q4 | 0.002 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.042 | 0.000 | 0.000 | 0.000 | 0.000 | 0.756 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.012 | 0.046 | 0.046 | 0.048 | 0.018 | 0.018 | 0.002 |
| Anxious_SkinThermo_Q3 | 0.002 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.044 | 0.000 | 0.000 | 0.000 | 0.000 | 0.754 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.012 | 0.046 | 0.046 | 0.048 | 0.018 | 0.018 | 0.002 |
| Sexual_SkinThermo_Q3 | 0.000 | 0.014 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.326 | 0.000 | 0.010 | 0.000 | 0.000 | 0.330 | 0.222 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.024 | 0.032 | 0.030 | 0.010 | 0.000 | 0.000 | 0.000 |
| Sexual_Respiratory_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.994 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sexual_Cardiac_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.994 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sexual_SkinThermo_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.012 | 0.960 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.008 | 0.010 | 0.002 | 0.004 | 0.000 | 0.000 |
| Sexual_SkinThermo_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.044 | 0.918 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.014 | 0.010 | 0.002 | 0.004 | 0.000 | 0.000 |
| Sexual_Gastric_Q2 | 0.002 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.200 | 0.000 | 0.000 | 0.000 | 0.000 | 0.036 | 0.660 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.016 | 0.038 | 0.022 | 0.008 | 0.000 | 0.000 | 0.000 |
| Sensitivity_ColonBladder_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.998 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_ColonBladder_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.998 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Nociception_ColonBladder_Q1 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.014 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.916 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.062 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 |
| Confusion_State_Q1 | 0.012 | 0.014 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.250 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.696 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.008 | 0.006 | 0.004 | 0.000 | 0.000 | 0.000 |
| Confusion_State_Q2 | 0.012 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.256 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.696 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.008 | 0.006 | 0.004 | 0.000 | 0.000 | 0.000 |
| Anxious_Cardiac_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.036 | 0.000 | 0.000 | 0.000 | 0.890 | 0.004 | 0.000 | 0.000 | 0.066 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_Respiratory_Q1 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.038 | 0.000 | 0.000 | 0.000 | 0.890 | 0.000 | 0.000 | 0.000 | 0.068 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_State_Q1 | 0.000 | 0.000 | 0.000 | 0.042 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.014 | 0.006 | 0.000 | 0.000 | 0.000 | 0.046 | 0.856 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.018 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_State_Q2 | 0.000 | 0.000 | 0.000 | 0.030 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.020 | 0.014 | 0.000 | 0.000 | 0.000 | 0.046 | 0.856 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.018 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_ColonBladder_Q1 | 0.000 | 0.256 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.008 | 0.176 | 0.000 | 0.000 | 0.000 | 0.000 | 0.098 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.398 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.038 | 0.008 | 0.006 | 0.000 | 0.000 | 0.000 |
| Anxious_Gastric_Q1 | 0.000 | 0.172 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.018 | 0.148 | 0.000 | 0.000 | 0.000 | 0.000 | 0.188 | 0.002 | 0.000 | 0.000 | 0.000 | 0.002 | 0.398 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.016 | 0.042 | 0.010 | 0.002 | 0.002 | 0.000 | 0.000 |
| Sensitivity_State_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.996 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_State_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.996 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_State_Q3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.130 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.868 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q7 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.996 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_State_Q5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.996 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Respiratory_Q7 | 0.000 | 0.000 | 0.072 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.924 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Respiratory_Q5 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.992 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q5 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.992 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Respiratory_Q6 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.000 | 0.974 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Genital_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.992 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Genital_Q3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.992 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Genital_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.992 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Confusion_Genital_Q1 | 0.034 | 0.122 | 0.000 | 0.000 | 0.000 | 0.000 | 0.014 | 0.000 | 0.016 | 0.296 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.472 | 0.000 | 0.000 | 0.002 | 0.014 | 0.016 | 0.006 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Gastric_Q2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Gastric_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Gastric_Q3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Gastric_Q6 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.992 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Gastric_Q5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.074 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.908 | 0.002 | 0.004 | 0.004 | 0.000 | 0.006 | 0.002 | 0.000 |
| Sensitivity_ColonBladder_Q3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.122 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.860 | 0.002 | 0.004 | 0.004 | 0.000 | 0.006 | 0.002 | 0.000 |
| Sensitivity_Gastric_Q4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.854 | 0.008 | 0.020 | 0.044 | 0.028 | 0.026 | 0.016 | 0.004 |
| Sensitivity_SkinThermo_Q5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.854 | 0.008 | 0.020 | 0.044 | 0.028 | 0.026 | 0.016 | 0.004 |
items <- select(items, -all_of(toremove2))
byfacet <- select(byfacet, -all_of(toremove2))
bymodality <- select(bymodality, -all_of(toremove2))
byrandom <- select(byrandom, -all_of(toremove2))ega2 <- bootEGA(
items,
EGA.type = "hierEGA",
model = "glasso", # BGGM
algorithm = "leiden", # walktrap
type="resampling",
plot.itemStability=FALSE,
typicalStructure=TRUE,
plot.typicalGraph=FALSE,
iter=500,
seed=3, ncores = 4)
save(ega2, file="models/ega2.RData")load("models/ega2.RData")
plot(itemStability(ega2, IS.plot=FALSE))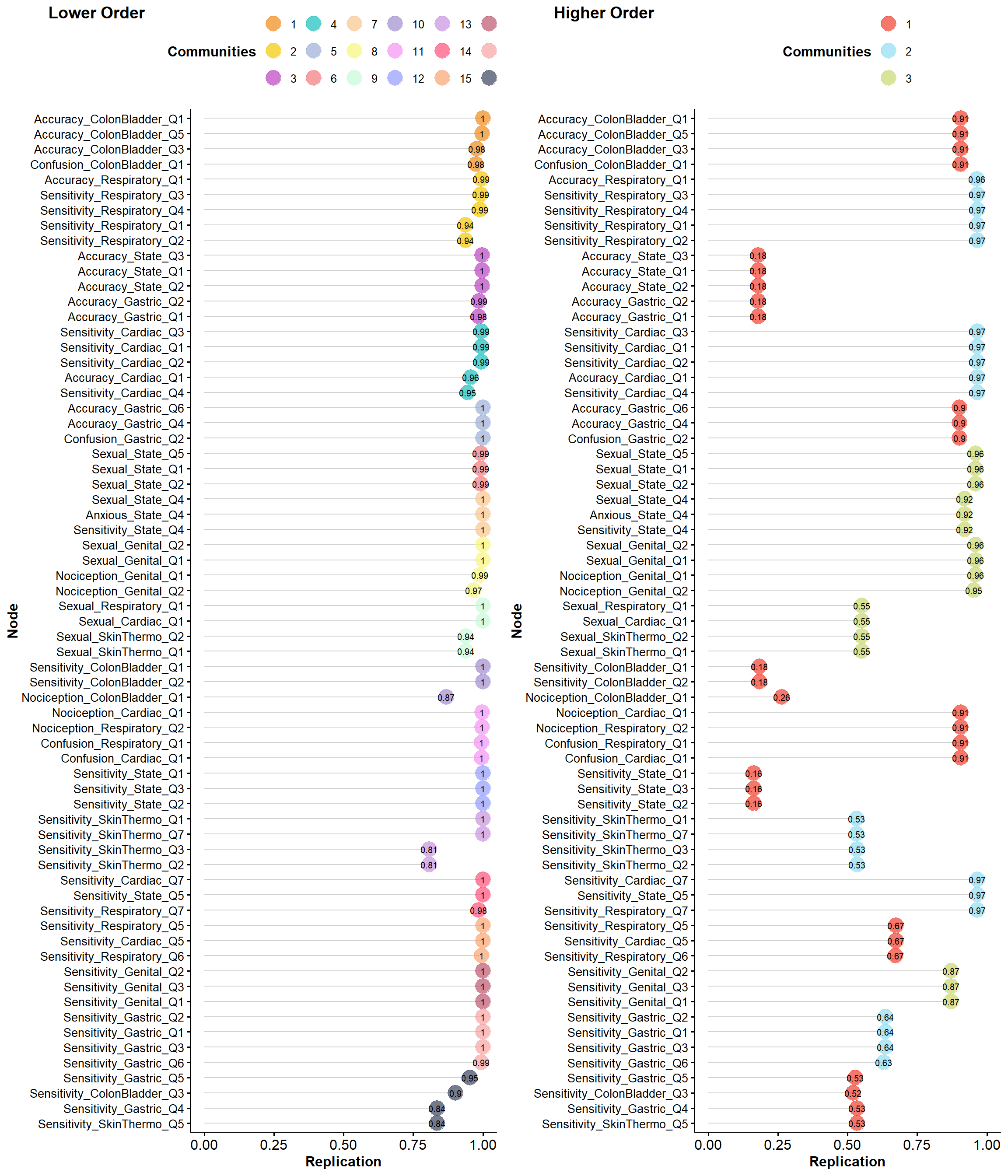
# itemstability_table <- make_loadingtable(itemstability$lower_order$item.stability$all.dimensions)
# toremove2 <- filter(itemstability_table, Max < 0.7)$Itemlower <- make_loadingtable(net.loads(ega2$EGA$lower_order)$std)
toremove3 <- lower$Item[!lower$Item %in% slice_max(lower, Max, n=3, by="Cluster")$Item]
lower |>
select(-Cluster, -Max) |>
gt::gt() |>
gt::data_color(columns=-Item,
method = "numeric",
palette = c("red", "white", "green"),
domain = c(-1, 1)) |>
gt::data_color(columns=Item, fn=\(x) ifelse(x %in% toremove3, "#FFEBEE", "white")) |>
gt::tab_header(
title = gt::md("**Item Loadings**"),
subtitle = "Node centrality"
) |>
gt::fmt_number(columns=-Item, decimals=3)| Item Loadings | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Node centrality | ||||||||||||||||||
| Item | C01 | C02 | C03 | C04 | C05 | C06 | C07 | C08 | C09 | C10 | C11 | C12 | C13 | C14 | C15 | C16 | C17 | C18 |
| Accuracy_ColonBladder_Q5 | 0.484 | 0.000 | 0.000 | 0.000 | 0.045 | 0.000 | 0.023 | 0.001 | 0.000 | 0.022 | 0.088 | 0.000 | 0.047 | 0.000 | 0.000 | 0.000 | 0.000 | 0.015 |
| Accuracy_ColonBladder_Q1 | 0.450 | 0.000 | −0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.039 | 0.013 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Confusion_ColonBladder_Q1 | 0.433 | 0.000 | −0.017 | 0.000 | 0.068 | −0.010 | −0.029 | 0.000 | 0.000 | 0.021 | 0.146 | −0.062 | 0.000 | 0.000 | 0.028 | 0.000 | 0.000 | 0.000 |
| Accuracy_ColonBladder_Q3 | 0.356 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | −0.007 | 0.000 | 0.028 | 0.029 | 0.025 | 0.000 | 0.000 | 0.000 | 0.035 | 0.009 | 0.028 | 0.035 |
| Sensitivity_Respiratory_Q3 | 0.000 | 0.501 | 0.000 | 0.096 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.016 | 0.052 | 0.047 | 0.000 | 0.003 | 0.010 | 0.000 |
| Sensitivity_Respiratory_Q1 | 0.000 | 0.469 | 0.000 | 0.113 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.021 | 0.000 | 0.000 | 0.081 | 0.005 | 0.000 | 0.027 | 0.000 |
| Sensitivity_Respiratory_Q4 | 0.000 | 0.446 | 0.000 | 0.127 | 0.000 | 0.000 | −0.026 | 0.000 | 0.006 | 0.000 | 0.000 | 0.035 | 0.000 | 0.000 | 0.000 | 0.043 | 0.048 | 0.000 |
| Sensitivity_Respiratory_Q2 | 0.000 | 0.352 | 0.000 | 0.125 | 0.000 | 0.000 | 0.000 | 0.000 | 0.034 | 0.000 | 0.078 | 0.000 | 0.000 | 0.092 | 0.000 | 0.000 | 0.035 | 0.000 |
| Accuracy_Respiratory_Q1 | 0.000 | 0.300 | 0.067 | 0.069 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.024 | −0.003 | 0.054 | 0.055 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Accuracy_State_Q2 | 0.000 | 0.002 | 0.544 | 0.000 | 0.000 | 0.042 | 0.000 | 0.000 | 0.000 | 0.015 | −0.002 | 0.019 | 0.022 | 0.019 | 0.007 | 0.000 | 0.000 | 0.000 |
| Accuracy_Gastric_Q2 | −0.034 | 0.002 | 0.439 | 0.012 | 0.000 | 0.009 | 0.000 | 0.008 | 0.000 | 0.016 | 0.000 | 0.042 | 0.000 | 0.007 | 0.000 | 0.000 | 0.001 | 0.008 |
| Accuracy_Gastric_Q1 | 0.000 | 0.000 | 0.408 | 0.000 | −0.011 | 0.000 | 0.000 | 0.000 | 0.000 | 0.035 | −0.021 | 0.029 | 0.043 | 0.000 | 0.007 | 0.032 | 0.005 | 0.050 |
| Accuracy_State_Q1 | 0.000 | 0.057 | 0.301 | 0.000 | 0.000 | 0.046 | 0.000 | 0.000 | 0.000 | 0.005 | 0.000 | 0.030 | 0.027 | 0.000 | 0.000 | 0.000 | 0.013 | 0.000 |
| Accuracy_State_Q3 | 0.000 | 0.004 | 0.288 | 0.000 | 0.000 | 0.030 | 0.018 | 0.057 | 0.000 | 0.034 | 0.000 | 0.040 | 0.064 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_Cardiac_Q3 | 0.000 | 0.078 | 0.000 | 0.533 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.046 | 0.000 | 0.011 | 0.041 | −0.019 | 0.000 | 0.025 | 0.000 |
| Sensitivity_Cardiac_Q2 | 0.000 | 0.110 | 0.000 | 0.527 | 0.000 | 0.000 | 0.000 | −0.020 | 0.000 | 0.000 | 0.020 | 0.000 | 0.000 | 0.082 | −0.022 | 0.000 | 0.000 | 0.002 |
| Sensitivity_Cardiac_Q1 | 0.000 | 0.106 | 0.000 | 0.438 | 0.007 | 0.000 | 0.000 | 0.000 | 0.007 | 0.000 | 0.069 | 0.000 | 0.017 | 0.018 | 0.000 | 0.028 | 0.016 | 0.005 |
| Accuracy_Cardiac_Q1 | 0.000 | 0.115 | 0.012 | 0.303 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.000 | 0.047 | 0.043 | 0.000 | 0.004 | 0.000 | 0.000 | 0.006 |
| Sensitivity_Cardiac_Q4 | 0.000 | 0.121 | 0.000 | 0.277 | 0.000 | 0.000 | 0.000 | −0.017 | 0.003 | 0.000 | 0.030 | 0.014 | 0.000 | 0.022 | −0.060 | 0.075 | 0.046 | 0.017 |
| Accuracy_Gastric_Q6 | 0.000 | 0.000 | 0.000 | 0.004 | 0.551 | 0.000 | 0.000 | 0.000 | 0.014 | 0.000 | 0.008 | 0.028 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Confusion_Gastric_Q2 | 0.094 | 0.000 | 0.000 | 0.002 | 0.398 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.040 | 0.000 | 0.000 | 0.000 | 0.009 | 0.000 | 0.000 | 0.008 |
| Accuracy_Gastric_Q4 | 0.000 | 0.000 | −0.008 | 0.000 | 0.332 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.029 | 0.000 | 0.000 | 0.000 | 0.030 | −0.012 | −0.014 | 0.000 |
| Sexual_State_Q2 | 0.000 | 0.000 | 0.018 | 0.000 | 0.000 | 0.538 | 0.036 | 0.115 | 0.021 | 0.027 | 0.000 | 0.012 | 0.000 | 0.000 | 0.000 | 0.034 | 0.000 | 0.000 |
| Sexual_State_Q5 | 0.000 | 0.000 | 0.071 | 0.000 | 0.000 | 0.468 | 0.021 | 0.092 | 0.028 | 0.000 | −0.003 | 0.071 | 0.000 | 0.002 | 0.014 | 0.043 | 0.000 | 0.000 |
| Sexual_State_Q1 | −0.010 | 0.000 | 0.032 | 0.000 | 0.000 | 0.451 | 0.096 | 0.103 | 0.000 | 0.009 | −0.058 | 0.049 | 0.000 | 0.000 | 0.000 | 0.058 | 0.000 | −0.014 |
| Sensitivity_State_Q4 | −0.006 | 0.000 | 0.017 | 0.000 | 0.000 | 0.003 | 0.500 | 0.018 | 0.020 | 0.014 | 0.000 | 0.123 | 0.028 | 0.014 | 0.000 | 0.000 | 0.003 | 0.000 |
| Sexual_State_Q4 | 0.022 | −0.023 | 0.000 | 0.000 | 0.000 | 0.144 | 0.458 | 0.093 | 0.023 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Anxious_State_Q4 | −0.029 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.456 | 0.028 | 0.000 | 0.000 | 0.000 | 0.000 | 0.041 | 0.026 | 0.000 | 0.000 | 0.000 | 0.000 |
| Nociception_Genital_Q1 | 0.000 | 0.000 | 0.000 | −0.015 | 0.000 | 0.063 | 0.000 | 0.566 | 0.005 | 0.009 | 0.000 | 0.000 | 0.025 | 0.000 | 0.000 | 0.099 | 0.000 | 0.000 |
| Sexual_Genital_Q2 | 0.000 | 0.000 | 0.000 | −0.019 | 0.000 | 0.100 | 0.030 | 0.466 | 0.051 | 0.030 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.000 | 0.000 |
| Sexual_Genital_Q1 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.138 | 0.095 | 0.396 | 0.089 | 0.003 | −0.004 | 0.000 | 0.012 | 0.000 | 0.008 | 0.084 | 0.000 | 0.000 |
| Nociception_Genital_Q2 | 0.001 | 0.000 | 0.061 | 0.000 | 0.000 | 0.010 | 0.018 | 0.241 | 0.000 | 0.104 | 0.000 | 0.000 | 0.011 | 0.000 | 0.002 | 0.048 | 0.000 | 0.004 |
| Sexual_SkinThermo_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.005 | 0.000 | 0.000 | 0.026 | 0.496 | 0.000 | 0.000 | 0.000 | 0.049 | 0.007 | 0.000 | 0.000 | 0.006 | 0.000 |
| Sexual_Respiratory_Q1 | 0.000 | 0.033 | 0.000 | 0.000 | 0.012 | 0.022 | 0.026 | 0.035 | 0.495 | 0.033 | 0.000 | 0.000 | 0.004 | 0.001 | 0.000 | 0.000 | 0.000 | 0.005 |
| Sexual_Cardiac_Q1 | 0.000 | 0.000 | 0.000 | 0.007 | 0.000 | 0.030 | 0.021 | 0.093 | 0.479 | 0.000 | 0.000 | 0.000 | 0.000 | 0.095 | 0.019 | 0.000 | 0.000 | 0.000 |
| Sexual_SkinThermo_Q2 | 0.029 | 0.006 | 0.000 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.306 | 0.000 | 0.008 | 0.000 | 0.054 | 0.000 | 0.069 | 0.014 | 0.004 | 0.035 |
| Sensitivity_ColonBladder_Q2 | 0.000 | 0.000 | 0.039 | 0.000 | 0.000 | 0.024 | 0.013 | 0.058 | 0.000 | 0.584 | 0.000 | 0.011 | 0.000 | 0.037 | 0.000 | 0.081 | 0.020 | 0.093 |
| Sensitivity_ColonBladder_Q1 | 0.000 | 0.020 | 0.047 | 0.009 | 0.000 | 0.008 | 0.000 | 0.056 | 0.000 | 0.493 | 0.000 | 0.036 | 0.026 | 0.026 | 0.000 | 0.052 | 0.039 | 0.043 |
| Nociception_ColonBladder_Q1 | 0.099 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.019 | 0.029 | 0.281 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.022 | 0.000 | 0.124 |
| Nociception_Respiratory_Q2 | 0.035 | 0.048 | −0.021 | 0.000 | 0.011 | −0.025 | 0.000 | 0.000 | 0.000 | 0.000 | 0.527 | −0.025 | 0.000 | 0.000 | 0.000 | −0.007 | 0.000 | 0.000 |
| Confusion_Respiratory_Q1 | 0.094 | 0.048 | 0.000 | 0.000 | 0.067 | −0.013 | 0.000 | 0.000 | 0.008 | 0.000 | 0.437 | −0.001 | 0.011 | 0.054 | 0.087 | 0.000 | 0.000 | 0.000 |
| Confusion_Cardiac_Q1 | 0.102 | 0.000 | 0.000 | 0.089 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.417 | 0.000 | 0.001 | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 |
| Nociception_Cardiac_Q1 | 0.049 | −0.003 | −0.002 | 0.069 | 0.000 | −0.027 | 0.000 | −0.004 | 0.001 | 0.000 | 0.388 | 0.000 | 0.023 | 0.000 | 0.001 | 0.000 | 0.000 | 0.017 |
| Sensitivity_State_Q1 | −0.060 | 0.065 | 0.054 | 0.006 | 0.000 | 0.063 | 0.060 | 0.000 | 0.000 | 0.000 | −0.016 | 0.589 | 0.019 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Sensitivity_State_Q2 | 0.000 | 0.002 | 0.044 | 0.022 | 0.000 | 0.063 | 0.055 | 0.000 | 0.000 | 0.051 | −0.006 | 0.526 | 0.005 | 0.017 | 0.000 | 0.005 | 0.035 | 0.004 |
| Sensitivity_State_Q3 | 0.000 | 0.028 | 0.052 | 0.027 | 0.033 | 0.002 | 0.011 | 0.000 | 0.000 | 0.000 | −0.003 | 0.325 | 0.000 | 0.000 | 0.036 | 0.001 | 0.000 | 0.000 |
| Sensitivity_SkinThermo_Q1 | 0.000 | 0.000 | 0.028 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.557 | 0.007 | 0.024 | 0.000 | 0.036 | 0.033 |
| Sensitivity_SkinThermo_Q7 | 0.047 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.021 | 0.000 | 0.012 | 0.000 | 0.035 | 0.000 | 0.439 | 0.000 | 0.003 | 0.000 | 0.000 | 0.006 |
| Sensitivity_SkinThermo_Q2 | 0.000 | 0.027 | 0.090 | 0.049 | 0.000 | 0.000 | 0.051 | 0.050 | 0.015 | 0.000 | 0.000 | 0.005 | 0.392 | 0.000 | 0.005 | 0.018 | 0.040 | 0.000 |
| Sensitivity_SkinThermo_Q3 | 0.000 | 0.075 | 0.036 | 0.012 | 0.000 | 0.000 | 0.004 | 0.000 | 0.077 | 0.029 | 0.000 | 0.020 | 0.346 | 0.000 | 0.000 | 0.021 | 0.041 | 0.045 |
| Sensitivity_Cardiac_Q7 | 0.000 | 0.000 | 0.000 | 0.118 | 0.000 | 0.000 | 0.000 | 0.000 | 0.079 | 0.000 | 0.000 | 0.000 | 0.000 | 0.562 | 0.000 | 0.000 | 0.000 | 0.006 |
| Sensitivity_Respiratory_Q7 | 0.000 | 0.200 | 0.000 | 0.013 | 0.000 | 0.000 | 0.000 | 0.000 | 0.007 | 0.000 | 0.050 | 0.000 | 0.007 | 0.454 | 0.037 | 0.000 | 0.000 | 0.030 |
| Sensitivity_State_Q5 | 0.000 | 0.000 | 0.024 | 0.016 | 0.000 | 0.001 | 0.041 | 0.000 | 0.011 | 0.072 | 0.000 | 0.017 | 0.000 | 0.425 | 0.000 | 0.020 | 0.060 | 0.001 |
| Sensitivity_Respiratory_Q5 | 0.051 | 0.000 | 0.000 | 0.000 | 0.037 | 0.000 | 0.000 | 0.000 | 0.046 | 0.000 | 0.022 | 0.000 | 0.000 | 0.002 | 0.502 | 0.008 | 0.000 | 0.065 |
| Sensitivity_Cardiac_Q5 | 0.000 | 0.000 | 0.011 | −0.076 | 0.000 | 0.011 | 0.000 | 0.001 | 0.008 | 0.000 | 0.000 | 0.006 | 0.020 | 0.000 | 0.432 | 0.008 | 0.000 | 0.045 |
| Sensitivity_Respiratory_Q6 | 0.000 | 0.004 | 0.000 | 0.003 | 0.000 | 0.000 | 0.000 | 0.007 | 0.015 | 0.000 | 0.051 | 0.024 | 0.005 | 0.029 | 0.311 | 0.003 | 0.000 | 0.031 |
| Sensitivity_Genital_Q1 | 0.000 | 0.000 | 0.000 | 0.000 | −0.014 | 0.067 | 0.000 | 0.154 | 0.002 | 0.044 | −0.003 | 0.000 | 0.000 | 0.015 | 0.013 | 0.591 | 0.000 | 0.000 |
| Sensitivity_Genital_Q2 | 0.000 | 0.000 | 0.031 | 0.026 | 0.000 | 0.070 | 0.000 | 0.074 | 0.000 | 0.078 | 0.000 | 0.005 | 0.038 | 0.006 | 0.000 | 0.465 | 0.026 | 0.014 |
| Sensitivity_Genital_Q3 | 0.009 | 0.043 | 0.000 | 0.070 | 0.000 | 0.000 | 0.000 | 0.013 | 0.011 | 0.051 | −0.004 | 0.001 | 0.000 | 0.000 | 0.010 | 0.418 | 0.024 | 0.042 |
| Sensitivity_Gastric_Q2 | 0.000 | 0.016 | 0.014 | 0.026 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.066 | 0.070 | 0.000 | 0.024 | 0.597 | 0.013 |
| Sensitivity_Gastric_Q1 | 0.032 | 0.028 | 0.000 | 0.016 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.006 | 0.000 | 0.021 | 0.042 | 0.000 | 0.000 | 0.000 | 0.563 | 0.000 |
| Sensitivity_Gastric_Q3 | 0.000 | 0.055 | 0.000 | 0.048 | −0.019 | 0.000 | 0.000 | 0.000 | 0.011 | 0.000 | 0.000 | 0.010 | 0.000 | 0.000 | 0.000 | 0.032 | 0.451 | 0.012 |
| Sensitivity_Gastric_Q6 | 0.000 | 0.027 | 0.007 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.000 | 0.068 | 0.000 | 0.009 | 0.021 | 0.000 | 0.000 | 0.000 | 0.313 | 0.168 |
| Sensitivity_SkinThermo_Q5 | 0.009 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.017 | 0.010 | 0.000 | 0.000 | 0.062 | 0.000 | 0.059 | 0.023 | 0.057 | 0.498 |
| Sensitivity_Gastric_Q4 | 0.033 | 0.000 | 0.006 | 0.020 | 0.010 | −0.013 | 0.000 | 0.000 | 0.015 | 0.010 | 0.016 | 0.004 | 0.017 | 0.007 | 0.050 | 0.025 | 0.041 | 0.438 |
| Sensitivity_Gastric_Q5 | 0.005 | 0.000 | 0.021 | 0.000 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.117 | 0.000 | 0.000 | 0.000 | 0.000 | 0.056 | 0.006 | 0.040 | 0.393 |
| Sensitivity_ColonBladder_Q3 | 0.000 | 0.000 | 0.022 | 0.007 | 0.000 | 0.000 | 0.000 | 0.000 | 0.005 | 0.139 | 0.000 | 0.000 | 0.000 | 0.029 | 0.002 | 0.000 | 0.027 | 0.288 |
items <- select(items, -all_of(toremove3))
byfacet <- select(byfacet, -all_of(toremove3))
bymodality <- select(bymodality, -all_of(toremove3))
byrandom <- select(byrandom, -all_of(toremove3))We kept the 3 items with the highest loading in their lower-level structure, renoving 13 items (Accuracy_ColonBladder_Q3, Sensitivity_Respiratory_Q2, Accuracy_Respiratory_Q1, Accuracy_State_Q1, Accuracy_State_Q3, Accuracy_Cardiac_Q1, Sensitivity_Cardiac_Q4, Nociception_Genital_Q2, Sexual_SkinThermo_Q2, Nociception_Cardiac_Q1, Sensitivity_SkinThermo_Q3, Sensitivity_Gastric_Q6, Sensitivity_ColonBladder_Q3).
ega3 <- bootEGA(
items,
EGA.type = "hierEGA",
model = "glasso", # BGGM
algorithm = "leiden", # walktrap
type="resampling",
plot.itemStability=FALSE,
typicalStructure=TRUE,
plot.typicalGraph=TRUE,
iter=500,
seed=3, ncores = 4)
save(ega3, file="models/ega3.RData")load("models/ega3.RData")
plot(itemStability(ega3))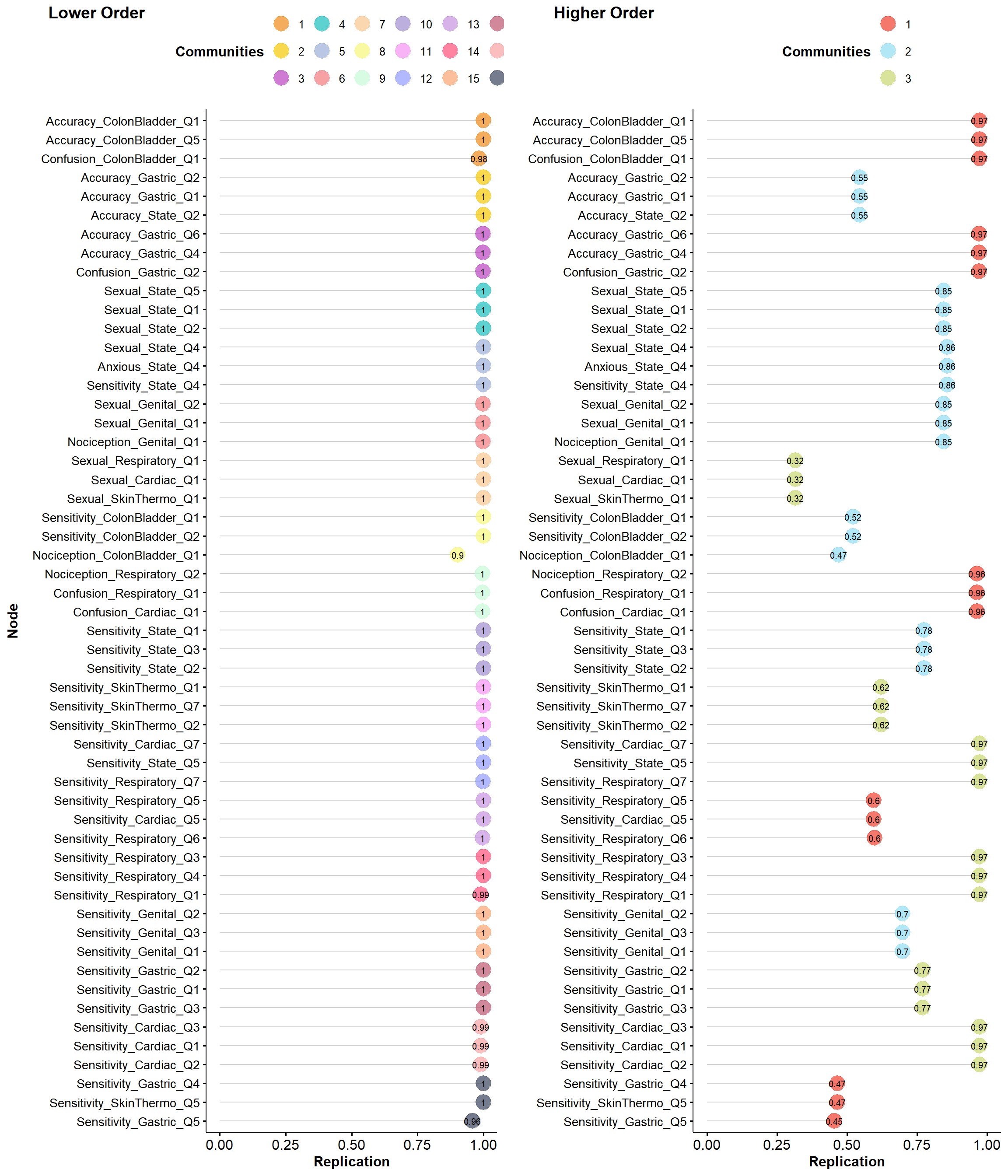
dimensions <- list(
C01 = "UrIn", # Urointestinal Inaccuracy
C09 = "CaCo", # Cardiorespiratory Confusion
C13 = "CaNo", # Cardiorespiratory Noticing
C18 = "Olfa", # Olfactory Compensation
C03 = "Sati", # Satiety Noticing
C04 = "SexA", # Sexual Arousal Awareness
C06 = "SexS", # Sexual Arousal Sensitivity
C15 = "SexO", # Sexual Organs Sensitivity
C08 = "UrSe", # Urointestinal Sensitivity
C10 = "RelA", # Relaxation Awareness
C05 = "StaS", # State Specificity
C02 = "ExAc", # Expulsion Accuracy
C17 = "Card", # Cardioception
C14 = "Resp", # Respiroception
C12 = "Sign", # Signalling
C16 = "Gast", # Gastroception
C11 = "Derm", # Dermatoception
C07 = "SexC" # Sexual Arousal Changes
)
higher <- make_loadingtable(t(net.loads(ega3$EGA$higher_order, loading.method="revised")$std))
cluster_order <- names(higher)[!names(higher) %in% c("Item", "Cluster", "Max")]
lower <- make_loadingtable(net.loads(ega3$EGA$lower_order, loading.method="revised")$std) |>
select(names(higher)) |>
mutate(Cluster = fct_relevel(Cluster, cluster_order)) |>
arrange(Cluster, desc(Max))
higher$Item <- paste0("M", higher$Item)
higher$Label <- c("Interoceptive Deficit", "Interoceptive Awareness", "Interoceptive Sensitivity")
lower <- mutate(lower, Label = labels[Item], ID= Item, Item = 1:n())
tab1 <- rbind(higher, select(lower, -ID)) |>
datawizard::data_relocate("Label", after = "Item") |>
select(-Cluster, -Max) |>
# datawizard::data_rename(dimensions) |> # https://github.com/easystats/datawizard/issues/576
datawizard::data_rename(names(dimensions), dimensions) |>
mutate(`|` = "") |>
gt::gt() |>
gt::tab_row_group(
label = "Items",
rows = 1:(nrow(lower) + nrow(higher))
) |>
gt::tab_row_group(
label = "Metaclusters",
rows = 1:nrow(higher)
) |>
gt::data_color(columns=-Item,
method = "numeric",
palette = c("red", "white", "green"),
domain = c(-1, 1)) |>
gt::tab_style(
style = gt::cell_text(size="small", style="italic"),
locations = gt::cells_body(columns="Label", rows=c(4:57))
) |>
gt::tab_style(
style = list(gt::cell_text(weight="bold"),
gt::cell_fill(color = "#F5F5F5")),
locations = list(
gt::cells_row_groups(groups = "Items"),
gt::cells_row_groups(groups = "Metaclusters")
)
) |>
# Metacluster 1
gt::tab_style(
style = gt::cell_fill(color = "#B39DDB"),
locations = list(
gt::cells_column_labels(columns = 3),
gt::cells_body(columns="Item", rows=c(4:6))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#9FA8DA"),
locations = list(
gt::cells_column_labels(columns = 4),
gt::cells_body(columns="Item", rows=c(7:9))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#90CAF9"),
locations = list(
gt::cells_column_labels(columns = 5),
gt::cells_body(columns="Item", rows=c(1, 10:12))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#81D4FA"),
locations = list(
gt::cells_column_labels(columns = 6),
gt::cells_body(columns="Item", rows=c(13:15))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#80DEEA"),
locations = list(
gt::cells_column_labels(columns = 7),
gt::cells_body(columns="Item", rows=c(16:18))
)
) |>
# Metacluster 2
gt::tab_style(
style = gt::cell_fill(color = "#F3F29D"),
locations = list(
gt::cells_column_labels(columns = 8),
gt::cells_body(columns="Item", rows=c(19:21))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#E6EE9C"),
locations = list(
gt::cells_column_labels(columns = 9),
gt::cells_body(columns="Item", rows=c(22:24))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#D6E8A1"),
locations = list(
gt::cells_column_labels(columns = 10),
gt::cells_body(columns="Item", rows=c(25:27))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#C5E1A5"),
locations = list(
gt::cells_column_labels(columns = 11),
gt::cells_body(columns="Item", rows=c(2, 28:30))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#B5DCA6"),
locations = list(
gt::cells_column_labels(columns = 12),
gt::cells_body(columns="Item", rows=c(31:33))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#A5D6A7"),
locations = list(
gt::cells_column_labels(columns = 13),
gt::cells_body(columns="Item", rows=c(34:36))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#93D1B6"),
locations = list(
gt::cells_column_labels(columns = 14),
gt::cells_body(columns="Item", rows=c(37:39))
)
) |>
# Metacluster 3
gt::tab_style(
style = gt::cell_fill(color = "#CE93D8"),
locations = list(
gt::cells_column_labels(columns = 15),
gt::cells_body(columns="Item", rows=c(40:42))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#F48FB1"),
locations = list(
gt::cells_column_labels(columns = 16),
gt::cells_body(columns="Item", rows=c(43:45))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#EF9A9A"),
locations = list(
gt::cells_column_labels(columns = 17),
gt::cells_body(columns="Item", rows=c(3, 46:48))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#FFAB91"),
locations = list(
gt::cells_column_labels(columns = 18),
gt::cells_body(columns="Item", rows=c(49:51))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#FFCC80"),
locations = list(
gt::cells_column_labels(columns = 19),
gt::cells_body(columns="Item", rows=c(52:54))
)
) |>
gt::tab_style(
style = gt::cell_fill(color = "#FFE082"),
locations = list(
gt::cells_column_labels(columns = 20),
gt::cells_body(columns="Item", rows=c(55:57))
)
) |>
gt::fmt_number(columns=-Item, decimals=2) |>
gt::tab_header(
title = gt::md("**Item Loadings**"),
subtitle = "Node centrality"
) |>
gt::opt_vertical_padding(scale = 0.7) |>
gt::opt_horizontal_padding(scale = 0.5) |>
# gt::tab_options(table.width = gt::pct(100)) |>
gt::cols_width(Label ~ gt::pct(66),
`|` ~ gt::pct(1))
gt::gtsave(gt::cols_hide(tab1, `|`), "table1.html", inline_css = TRUE) # Save it and reload it otherwise formatting not ideal
Sys.setenv(CHROMOTE_CHROME = "C:\\Program Files (x86)\\Microsoft\\Edge\\Application\\msedge.exe")
gt::gtsave(tab1, "table1.png", vwidth=1660, expand=0, quiet=TRUE)# REMOVE ONCE https://github.com/hfgolino/EGAnet/pull/167 is merged
convert2tidygraph <- function(EGA.object)
{
if("bootEGA" %in% class(EGA.object)) {
# Bootstrapped EGA ---
if(!"typicalGraph" %in% names(EGA.object)) {
stop("The bootEGA object must contain a typicalGraph object. Set `typicalStructure=TRUE`.")
}
if("lower_order" %in% names(EGA.object$typicalGraph)) {
# Hierarchical EGA ---
lower_nodes <- .convert2tidygraph_nodes(EGA.object$typicalGraph$lower_order$wc)
lower_nodes$dimension <- paste0("L", lower_nodes$dimension) # Discriminate from higher
lower_nodes$level <- "Lower"
higher_nodes <- .convert2tidygraph_nodes(EGA.object$typicalGraph$higher_order$wc)
higher_nodes$name <- paste0("L", as.numeric(higher_nodes$name))
higher_nodes$dimension <- paste0("H", higher_nodes$dimension) # Discriminate from lower
higher_nodes$level <- "Higher"
nodes <- rbind(lower_nodes, higher_nodes)
lower_edges <- .convert2tidygraph_edges(EGA.object$typicalGraph$lower_order$graph)
higher_edges <- .convert2tidygraph_edges(EGA.object$typicalGraph$higher_order$graph)
higher_edges$from <- paste0("L", as.numeric(higher_edges$from))
higher_edges$to <- paste0("L", as.numeric(higher_edges$to))
edges <- rbind(lower_edges, higher_edges)
edges$type <- "real"
# Make edges from lower to higher order
for(i in 1:nrow(lower_nodes)) {
edges <- rbind(
edges,
data.frame(from = lower_nodes$name[i], to = lower_nodes$dimension[i], link = 1, type = "virtual")
)
}
} else {
# Non-hierarchical EGA ---
nodes <- .convert2tidygraph_nodes(EGA.object$typicalGraph$wc)
edges <- .convert2tidygraph_edges(EGA.object$typicalGraph$graph)
}
} else {
# Normal EGA ---
nodes <- .convert2tidygraph_nodes(EGA.object$wc)
edges <- .convert2tidygraph_edges(EGA.object$network)
}
# Return result
return(
list(
nodes = nodes,
edges = edges[edges$link != 0,]
)
)
}
#' @keywords internal
#' @noRd
.convert2tidygraph_nodes <- function(wc){
data.frame(
name = names(wc),
dimension = as.character(wc)
)
}
#' @keywords internal
#' @noRd
.convert2tidygraph_edges <- function(network){
data.frame(
from = rep(rownames(network), times = ncol(network)),
to = rep(colnames(network), each = nrow(network)),
link = as.vector(network)
)
}#| warning: false
graph <- convert2tidygraph(ega3)
replace_lowernames <- function(names, lower) {
rownames(lower) <- lower$ID
sprintf("%02d", as.numeric(lower[names, "Item"]))
}
replace_highernames <- function(names) {
x <- as.numeric(str_remove(names, "L"))
paste0("C", sprintf("%02d", x))
}
# Arrange lower nodes in circle
lower_nodes <- graph$nodes |>
filter(level == "Lower") |>
mutate(name = fct_relevel(name, as.character(lower$ID)),
ID = name) |>
arrange(name)
lower_edges <- graph$edges |>
filter(from %in% lower_nodes$name & to %in% lower_nodes$name)
lower_layout <- tidygraph::tbl_graph(lower_nodes, edges=lower_edges, directed=FALSE) |>
ggraph::create_layout(layout = 'circle') |>
as.data.frame()
rownames(lower_layout) <- lower_nodes$name
# Arrange higher nodes in circle
higher_nodes <- graph$nodes |>
filter(level == "Higher") |>
mutate(ID = name,
name = replace_highernames(name),
name = fct_relevel(name, names(select(higher, -Item, -Cluster,-Max,-Label)))) |>
arrange(name)
higher_edges <- graph$edges |>
filter(from %in% higher_nodes$name & to %in% higher_nodes$name)
higher_layout <- tidygraph::tbl_graph(higher_nodes, edges=higher_edges, directed=FALSE) |>
ggraph::create_layout(layout = 'circle') |>
as.data.frame()
rownames(higher_layout) <- higher_nodes$ID
# Put circles together
layout <- tidygraph::tbl_graph(graph$nodes, edges=graph$edges, directed=FALSE) |>
ggraph::create_layout(layout = 'linear', circular = TRUE)
layout[layout$level == "Lower", "x"] <- lower_layout[layout[layout$level == "Lower", ]$name, "x"]
layout[layout$level == "Lower", "y"] <- lower_layout[layout[layout$level == "Lower", ]$name, "y"]
layout[layout$level == "Higher", "x"] <- higher_layout[layout[layout$level == "Higher", ]$name, "x"] * 0.5
layout[layout$level == "Higher", "y"] <- higher_layout[layout[layout$level == "Higher", ]$name, "y"] * 0.5
layout[layout$level == "Lower", "name"] <- replace_lowernames(layout[layout$level == "Lower", ]$name, lower)
layout[layout$level == "Higher", "name"] <- replace_highernames(layout[layout$level == "Higher", ]$name)
# Final renaming
layout[layout$level == "Higher", "name"] <- unlist(dimensions)[layout[layout$level == "Higher", ]$name]
layout[layout$level == "Higher", "dimension"] <- layout[layout$level == "Higher", ]$dimension |>
str_replace("H1", "Deficit") |>
str_replace("H2", "Awareness") |>
str_replace("H3", "Sensitivity")
# layout$dimmension_label <- layout$dimension
# layout[layout$level == "Higher", "dimmension_label"] <- NA
# layout[layout$level == "Higher", "dimension"] <- layout[layout$level == "Higher", "dimension"]
ggraph(layout) +
geom_edge_link(aes(edge_alpha=as.numeric(type == "virtual"), edge_width = abs(link)), edge_width=0.2, color="grey") +
geom_edge_arc(aes(edge_color = link, edge_width = abs(link), edge_alpha=as.numeric(type == "real"))) +
geom_node_point(aes(color=dimension, shape=level), size=12) +
geom_node_text(aes(label=name), fontface="bold") +
# geom_node_text(aes(label = dimmension_label, angle = node_angle(x, y), hjust = 'outward')) +
scale_edge_alpha(range = c(0, 0.9), guide="none") +
scale_edge_width(range = c(0.2, 7), guide="none") +
scale_edge_colour_gradientn(colors=c("#E53935", "#E53935", "white", "#43A047", "#43A047"),
values=c(-1, -0.5, 0, 0.5, 1), guide="none") +
scale_shape_manual(values=c("Higher"="square", "Lower"="circle"), guide="none") +
scale_color_manual(values=c(
"Deficit"="#90CAF9", "Awareness"="#C5E1A5", "Sensitivity"="#EF9A9A",
"L1"="#B39DDB", "L9"="#9FA8DA", "L13"="#90CAF9", "L18"="#81D4FA", "L3"="#80DEEA",
"L4"="#F3F29D", "L6"="#E6EE9C", "L15"="#D6E8A1", "L8"="#C5E1A5", "L10"="#B5DCA6", "L5"="#A5D6A7", "L2"="#93D1B6",
"L17"="#CE93D8", "L14"="#F48FB1", "L12"="#EF9A9A", "L16"="#FFAB91", "L11"="#FFCC80", "L7"="#FFE082"),
breaks=c("Deficit", "Awareness", "Sensitivity")) +
theme_void() +
theme(legend.title = element_text(face="bold")) +
labs(color="Metaclusters")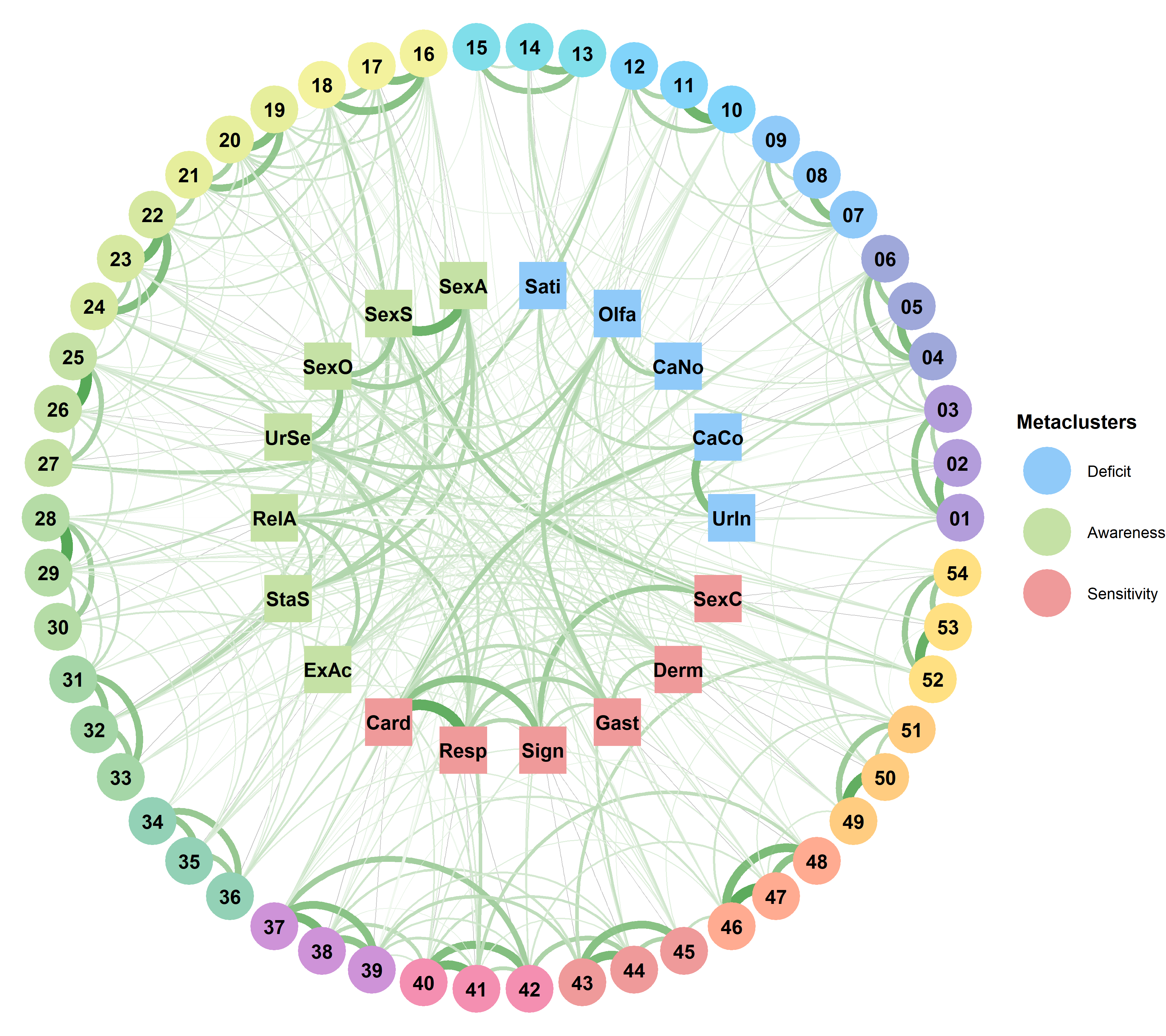
# EGAnet::EGM(items[, 1:60])
EGAnet::EGM.compare(items, model = "glasso", algorithm = "leiden")
rez <- EGAnet::network.compare(byfacet, bymodality, model = "glasso", seed=3)
plot(rez)# Modelbased scores
scores <- EGAnet::net.scores(
items[names(ega3$typicalGraph$lower_order$wc)],
A=ega3$typicalGraph$lower_order$graph,
wc=ega3$typicalGraph$lower_order$wc)$scores$std.scores |>
as.data.frame()
names(scores) <- paste0("C", names(scores))
# Empirical scores
clusters <- data.frame(Item = names(ega3$typicalGraph$lower_order$wc),
Cluster = ega3$typicalGraph$lower_order$wc) |>
mutate(Cluster = paste0("E", sprintf("%02d", Cluster)))
empirical <- data.frame(Participant=df$Participant)
for(c in unique(clusters$Cluster)) {
s <- rowMeans(items[clusters[clusters$Cluster==c, "Item"]])
empirical <- cbind(empirical, setNames(data.frame(x = s), c))
}
correlation::correlation(empirical, scores) |>
ggplot(aes(x=Parameter1, y=Parameter2, fill=r)) +
geom_tile() +
geom_text(aes(label=sprintf("%.2f", r)), size=3) +
scale_fill_gradient2(low="blue", high="red", mid="white", midpoint=0) +
labs(x="Empirical Scores", y="Model Scores") +
theme_minimal()